Dentatorubral–pallidoluysian atrophy
Dentatorubral–pallidoluysian atrophy (DRPLA) is an autosomal dominant spinocerebellar degeneration caused by an expansion of a CAG repeat encoding a polyglutamine tract in the atrophin-1 protein.[1] It is also known as Haw River Syndrome and Naito–Oyanagi disease. Although this condition was perhaps first described by Smith et al. in 1958, and several sporadic cases have been reported from Western countries, this disorder seems to be very rare except in Japan.
| Dentatorubral–pallidoluysian atrophy | |
|---|---|
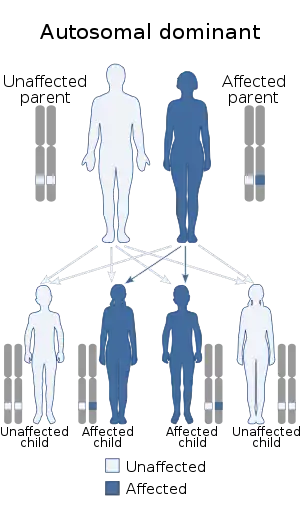 | |
| Dentatorubral–pallidoluysian atrophy is inherited in an autosomal dominant manner. | |
| Specialty | Neurology |
There are at least eight neurodegenerative diseases that are caused by expanded CAG repeats encoding polyglutamine (polyQ) stretches (see: Trinucleotide repeat disorder). The expanded CAG repeats create an adverse gain-of-function mutation in the gene products. Of these diseases, DRPLA is most similar to Huntington's disease.
Signs and symptoms
DRPLA can be juvenile-onset (<20 years), early adult-onset (20–40 years), or late adult-onset (>40 years). Late adult-onset DRPLA is characterized by ataxia, choreoathetosis and dementia. Early adult-onset DRPLA also includes seizures and myoclonus. Juvenile-onset DRPLA presents with ataxia and symptoms consistent with progressive myoclonus epilepsy [2] (myoclonus, multiple seizure types and dementia). Other symptoms that have been described include cervical dystonia,[3] corneal endothelial degeneration[4] autism, and surgery-resistant obstructive sleep apnea.[5]
Genetics
The human genome contains two atrophin genes; DRPLA has been correlated to the expansion of the polyglutamine region of the atrophin-1 gene on chromosome 12p13.3.[6] A normal number of CAG repeats in the atrophin-1 gene is 7–34, affected individuals display 49–93 repeats. DRPLA displays anticipation (earlier age of onset for subsequent generations) and an inverse correlation between the size of the expanded CAG repeat and the age of symptom onset. Paternal transmission results in more prominent anticipation (26–29 years) than maternal transmission (14–15 years).[2]
Atrophin-1
Atrophin-1 (ATN1) encodes a hydrophilic 1184 amino acid protein with several repetitive motifs including a serine-rich region, a variable length polyglutamine tract, a polyproline tract, and a region of alternating acidic and basic residues. It contains a putative nuclear localization signal in the N-terminus of the protein and a putative nuclear export signal in the C-terminus.[7] ATN1 is ubiquitously expressed in all tissues, but proteolytically cleaved in neuronal cells. The function of ATN1 is not clear, however it is believed to be a transcriptional co-repressor. ATN1 and atrophin-2 can be co-immunoprecipitated, indicating that they may carry out some functions together in a molecular complex.[8] Atrophin-1 may be a dispensable or redundant protein as mice bred with a null allele for atrophin-1 produce viable and fertile offspring and show no compensatory upregulation of atrophin-2.[9]
Transgenic mouse models
Mouse models of DRPLA have been successfully generated, which demonstrate the same intergenerational instability and severe phenotype as human DRPLA.[10][11][12] The Schilling mice express full-length human atrophin-1 with 65 CAG repeats under transcriptional control of the mouse prion protein promoter. The mice demonstrated progressive ataxia, tremors, abnormal movements, seizures and premature death. Like in human brains, nuclear accumulation was demonstrated and occasional NIIs were visualised, but the NIIs did not stain for ubiquitin and no neuronal loss was seen.[13] The Sato mice harbored a single copy of full-length human atrophin-1 with 76 or 129 CAG repeats. The hemizygous transgenic offspring of the Q129 mice exhibited symptoms similar to juvenile-type DRPLA, such as myoclonus and seizures. Again, neuronal atrophy was noted, but no neuronal loss (until death). Diffuse accumulation in the nuclei began on post-natal day 4 and ubiquitinated NII formation was detectable at 9 weeks of age. No PML bodies were found to be associated with the NIIs, which were morphologically mildly altered from those seen in human neural cells.[13][14]
Pathology
DRPLA is characterized by marked, generalized brain atrophy and the accumulation of atrophin-1 with expanded glutamine stretches. Mutant atrophin-1 proteins have been found in neuronal intranuclear inclusions (NII) and diffusely accumulated in the neuronal nuclei. While the role of NIIs (pathologic or protective) is unclear, the diffuse accumulation of mutant protein is regarded as toxic.
Brain atrophy
There is significant reduction in CNS tissue throughout the brain and spinal cord, with brain weights of DRPLA patients often becoming less than 1000g.[15] In regions lacking obvious neuronal depletion, atrophy of the neuropil is noted. The globus pallidus (lateral greater than medial segment) and subthalamic nucleus demonstrate consistent neuronal loss and astrocytic gliosis. The dentate nucleus shows neuronal loss with the remaining atrophic neurons exhibiting grumose degeneration. In general, the pallidoluysian degeneration is more severe than the dentatorubral degeneration in juvenile-onset and the reverse is true for the late adult-onset.[13]
Transgenic DRPLA mice demonstrated several neuronal abnormalities including a reduction in the number and size of dendritic spines as well as in the area of perikarya and diameter of dendrites.[14] Spine morphology and density have been linked to learning and memory functions as well as epilepsy. The stubby-type spines seen in DRPLA mice are morphologically different from the thin and mushroom-type spines seen in Huntington’s mice.
Morphometric analysis of DRPLA mouse brains has shown a loss of normal inter-microtubule spacing in neuronal axons. The microtubules were relatively compacted, suggesting abnormalities in protein transport may play a role in neuronal degeneration.[14] In humans, atrophin-1 interacts with IRSp53, which interacts with Rho GTPases to regulate the organization of the actin cytoskeleton and the pathways that regulate lamellipodia and filopodia.[16]
Neuronal intranuclear inclusions
NIIs are not exclusive to DRPLA; they have been found in a variety of neurodegenerative disorders. In DRPLA, NIIs have been demonstrated in both neurons and glial cells in the striatum, pontine nuclei, inferior olive, cerebellar cortex and dentate nucleus,[17] though the incidence of neurons with NIIs is low, roughly 1-3%.[13]
In DRPLA, the NIIs are spherical, eosinophilic structures of various sizes. They are non-membrane-bound and are composed of both granular and filamentous structures. They are ubiquitinated and may be paired or in doublet form within the nucleus.[18]
NIIs are immunopositive for several transcription factors such as TATA binding protein (TBP), TBP-associated factor (TAFII130), Sp1, camp-responsive element-binding protein (CREB) and CREB-binding protein (CBP).[19][20] It has been proposed that recruitment of transcription factors into NIIs may induce transcriptional abnormalities that contribute to progressive neuronal degeneration.[13] Other polyQ disorders, such as Huntington’s and spinocerebellar ataxia (types 3 and 7), have been demonstrated to sequester some of the same transcriptions factors. That different gene products sequester the same transcription factors may contribute to the overlapping symptoms of genetically different diseases.[21]
NIIs have also been demonstrated to alter the distribution of the intranuclear structures, such as promyelocytic leukemia protein (PML) nuclear bodies. Although the role of PML bodies is unclear, they are believed to be involved in apoptosis. In neurons with NII, PML bodies in DRPLA patients form a shell or ring around the ubiquitinated core.[13][21] In similar polyQ diseases, the association of this PML shell has been shown to be size-dependent with larger NIIs being PML negative.[22][23] This has led to two models, one in which PML bodies represent sites for NII formation and a second in which PML bodies are involved in degradation and proteolysis of NIIs.[21]
Filementous, atrophin-1 positive, inclusions are also observed exclusively in the cytoplasm of the dentate nucleus, which are extremely similar to the inclusions observed in the motor neurons in amyotrophic lateral sclerosis.[24]
Diffuse accumulation in the nuclei
In DRPLA, diffuse accumulation of mutant ATN1 occurs far more extensively than NII formation. The extent and frequency of neurons showing the diffuse nuclear accumulations changes depending on CAG repeat length. It is believed that the diffuse nuclear accumulations contribute to the clinical features such as dementia and epilepsy.
ATN1 contains both a nuclear localization sequence and a nuclear export sequence. Cleavage of ATN1 to an N terminal fragment relieves ATN1 of its nuclear export signal and concentrates it in the nucleus. Increased nuclear concentrations have been demonstrated via transfection assay to enhance cellular toxicity.[7]
In both the juvenile and adult forms, regions in which more than 40% of neurons became immunoreactive to 1C2 (a monoclonal antibody against expanded polyglutamine stretches) included: the nucleus basalis of Meynert, large striatal neurons, globus pallidus, subthalamic nucleus, thalamic intralaminar nucleus, lateral geniculate body, oculomotor nucleus, red nucleus, substantia nigra, trigeminal motor nucleus, nucleus raphes pontis, pontine nuclei, vestibular nucleus, inferior olive and the cerebellar dentate nucleus. The juvenile type also shows reactivity in the cerebral cortex, hippocampal CA1 area, and the reticular formation of the brainstem.[13] Nuclei containing accumulations of mutant atrophin-1 are deformed with nuclear membrane indentations.[25]
Diagnosis
Diagnosis of DRPLA rests on positive family history, clinical findings, and genetic testing. Family history can be difficult to obtain if a relative was misdiagnosed, died young, or experiences late onset of symptoms.
Other diseases in the differential diagnosis of adult-onset DRPLA include Huntington's and the spinocerebellar ataxias. For juvenile-onset disease, familial essential myoclonus and epilepsy (FEME), Lafora, Unverricht-Lundborg, Neuroaxonal dystrophy, Gaucher's disease, Sialidosis, and Galactosialidosis should be considered.
Management
To quantify the extent of the disease, an MRI, EEG and neuropsychological testing are recommended. Seizures are treated with anticonvulsants and psychiatric disturbances with psychotropic medications. Physical therapy has also been recommended to maintain function as the condition progresses and occupational therapy to focus on activities of daily living, advice for carers and adaptation to the environment.
Epidemiology
The prevalence of DRPLA in the Japanese population is believed to be 2–7 in 1,000,000. DRPLA is observed relatively less frequently in other ethnic populations and an analysis of normal ATN1 alleles has demonstrated that CAG repeat lengths greater than 17 are significantly more frequent in the Japanese population.[26][27]
References
- Kanazawa I (June 1999). "Molecular pathology of dentatorubral–pallidoluysian atrophy". Philos. Trans. R. Soc. Lond. B Biol. Sci. 354 (1386): 1069–74. doi:10.1098/rstb.1999.0460. PMC 1692599. PMID 10434307.
- Tsuji, S. (1999). "Dentatorubral–pallidoluysian atrophy: Clinical features and molecular genetics". Adv Neurol. 79: 399–409. PMID 10514829.
- Hatano, T.; et al. (2003). "Cervical dystonia in dentatorubral–pallidoluysian atrophy". Acta Neurol Scand. 108 (4): 287–9. doi:10.1034/j.1600-0404.2003.00150.x. PMID 12956864.
- Ito, D.; et al. (2002). "Corneal endothelial degeneration in dentatorubral–pallidoluysian atrophy". Arch Neurol. 59 (2): 289–91. doi:10.1001/archneur.59.2.289. PMID 11843701.
- Licht D, Lynch D (2002). "Juvenile Dentatorubral–Pallidoluysian Atrophy: New Clinical Features". Pediatr Neurol. 26 (1): 51–4. doi:10.1016/S0887-8994(01)00346-0. PMID 11814736.
- Yazawa, I; et al. (1995). "Abnormal Gene Product Identified in Hereditary DRPLA Brain". Nat Genet. 10 (1): 99–103. doi:10.1038/ng0595-99. PMID 7647802.
- Nucifora, F; et al. (2003). "Nuclear localization of a Non-caspase Truncation Product of Atrophin-1, with an Expanded Polyglutamine Repeat, Increases Cellular Toxicity". J Biol Chem. 278 (15): 13047–55. doi:10.1074/jbc.M211224200. PMID 12464607.
- Zoltewicz, J; et al. (2004). "Atrophin-2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis". Development. 131 (1): 3–14. doi:10.1242/dev.00908. PMID 14645126.
- Shen, Y; et al. (2007). "Functional Architecture of Atrophins". J Biol Chem. 282 (7): 5037–44. doi:10.1074/jbc.M610274200. PMID 17150957.
- Sato, T; et al. (1999). "Transgenic mice harboring a full-length human mutant DRPLA gene exhibit age-dependent intergenerational and somatic instabilities of CAG repeats comparable with those in DRPLA patients". Hum Mol Genet. 8 (1): 99–106. doi:10.1093/hmg/8.1.99. PMID 9887337.
- Sato, T; et al. (1999). "Transgenic mice harboring a full-length human DRPLA gene with highly expanded CAG repeats exhibit severe disease phenotype". Am J Hum Genet. 65 (suppl): A30.
- Schilling, G; et al. (1999). "Nuclear accumulation of truncated atrophin-1 fragments in a transgenic mouse model of DRPLA". Neuron. 24 (1): 275–86. doi:10.1016/S0896-6273(00)80839-9. PMID 10677044.
- Yamada, M; et al. (2008). "CAG repeat disorder models and human neuropathology: similarities and differences". Acta Neuropathol. 115 (1): 71–86. doi:10.1007/s00401-007-0287-5. PMID 17786457.
- Sakai, K; et al. (2006). "Neuronal Atrophy and Synaptic Alteration in a Mouse Model of Dentatorubral–pallidoluysian Atrophy". Brain. 129 (Pt 9): 2353–62. doi:10.1093/brain/awl182. PMID 16891319.
- Naito H, Oyanagi S (1982). "Familial myoclonus epilepsy and choreoathetosis: hereditary dentatorubral–pallidoluysian atrophy". Neurology. 32 (8): 798–807. doi:10.1212/wnl.32.8.798. PMID 6808417.
- Mackie S, Aitken A (2005). "Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease". FEBS. 272 (16): 4202–10. doi:10.1111/j.1742-4658.2005.04832.x. PMID 16098201.
- Hayashi, Y; et al. (1998). "Hereditary dentatorubral–pallidoluysian atrophy: Detection of widespread ubiquitinated neuronal and glial intranuclear inclusions in the brain". Acta Neuropathol. 96 (6): 547–52. doi:10.1007/s004010050933. PMID 9845282.
- Yamada, M; et al. (2001). "Interaction between Neuronal Intranuclear Inclusions and Promyelocytic Leukemia Protein Nuclear and Coiled Bodies in CAG Repeat Diseases". Am J Pathol. 159 (5): 1785–95. doi:10.1016/S0002-9440(10)63025-8. PMC 1867069. PMID 11696439.
- Yamada, M; et al. (2001). "Widespread occurrence of intranuclear atrophin-1 accumulation in the central nervous system neurons of patients with dentatorubral–pallidoluysian atrophy". Ann Neurol. 49 (1): 14–23. doi:10.1002/1531-8249(200101)49:1<14::AID-ANA5>3.0.CO;2-X. PMID 11198291.
- Shimohata, T; et al. (2000). "Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription". Nat Genet. 26 (1): 29–36. doi:10.1038/79139. PMID 10973244.
- Woulfe, JM (2007). "Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress". Neuropathol Appl Neurobiol. 33 (1): 2–42. doi:10.1111/j.1365-2990.2006.00819.x. PMID 17239006.
- Takahashi-Fujigasaki, J; et al. (2006). "SUMOylation substrates in neuronal intranuclear inclusion disease". Neuropathol Appl Neurobiol. 32 (1): 92–100. doi:10.1111/j.1365-2990.2005.00705.x. PMID 16409557.
- Takahashi, J; et al. (2002). "Two populations of neuronal intranuclear inclusions in SCA7 differ in size and promyelocytic leukaemia protein content". Brain. 125 (7): 1534–43. doi:10.1093/brain/awf154. PMID 12077003.
- Yamada, M; et al. (2000). "Ubiquitinated filamentous inclusions in cerebellar dentate nucleus neurons in dentatorubral–pallidoluysian atrophy contain expanded polyglutamine stretches". Acta Neuropathol. 99 (6): 615–8. doi:10.1007/s004010051171. PMID 10867794.
- Takahashi, J; et al. (2001). "Neuronal nuclear alterations in dentatorubral–pallidoluysian atrophy: ultrastructural and morphometric studies of the cerebellar granule cells". Brain Res. 919 (1): 12–9. doi:10.1016/S0006-8993(01)02986-9. PMID 11689158.
- Burke, JR; et al. (1994). "Dentatorubral–pallidoluysian atrophy and Haw River Syndrome". Lancet. 344 (8938): 1711–2. doi:10.1016/S0140-6736(94)90497-9. PMID 7996992.
- Takano, H; et al. (1998). "Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations". Am J Hum Genet. 63 (4): 1060–6. doi:10.1086/302067. PMC 1377499. PMID 9758625.
External links
| Classification | |
|---|---|
| External resources |