Gene silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene.[1][2] Gene silencing can occur during either transcription or translation and is often used in research.[1][2] In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.
Gene silencing is often considered the same as gene knockdown.[3][4] When genes are silenced, their expression is reduced.[3][4] In contrast, when genes are knocked out, they are completely erased from the organism's genome and, thus, have no expression.[3][4] Gene silencing is considered a gene knockdown mechanism since the methods used to silence genes, such as RNAi, CRISPR, or siRNA, generally reduce the expression of a gene by at least 70% but do not completely eliminate it. Methods using gene silencing are often considered better than gene knockouts since they allow researchers to study essential genes that are required for the animal models to survive and cannot be removed. In addition, they provide a more complete view on the development of diseases since diseases are generally associated with genes that have a reduced expression.[3]
Types
Transcriptional
- Genomic Imprinting
- Paramutation
- Transposon silencing (or Histone Modifications)
- Transgene silencing
- Position effect
- RNA-directed DNA methylation
Post-transcriptional
Research methods
Antisense oligonucleotides
Antisense oligonucleotides were discovered in 1978 by Paul Zamecnik and Mary Stephenson.[5] Oligonucleotides, which are short nucleic acid fragments, bind to complementary target mRNA molecules when added to the cell.[5][6] These molecules can be composed of single-stranded DNA or RNA and are generally 13–25 nucleotides long.[6][7] The antisense oligonucleotides can affect gene expression in two ways: by using an RNase H-dependent mechanism or by using a steric blocking mechanism.[6][7] RNase H-dependent oligonucleotides cause the target mRNA molecules to be degraded, while steric-blocker oligonucleotides prevent translation of the mRNA molecule.[6][7] The majority of antisense drugs function through the RNase H-dependent mechanism, in which RNase H hydrolyzes the RNA strand of the DNA/RNA heteroduplex.[6][7] This mechanism is thought to be more efficient, resulting in an approximately 80% to 95% decrease in the protein and mRNA expression.[6]
Ribozymes
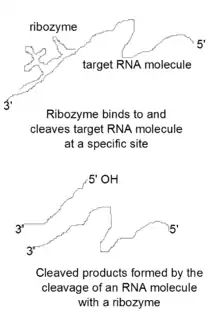
Ribozymes are catalytic RNA molecules used to inhibit gene expression. These molecules work by cleaving mRNA molecules, essentially silencing the genes that produced them. Sidney Altman and Thomas Cech first discovered catalytic RNA molecules, RNase P and group II intron ribozymes, in 1989 and won the Nobel Prize for their discovery.[8][9] Several types of ribozyme motifs exist, including hammerhead, hairpin, hepatitis delta virus, group I, group II, and RNase P ribozymes. Hammerhead, hairpin, and hepatitis delta virus (HDV) ribozyme motifs are generally found in viruses or viroid RNAs.[8] These motifs are able to self-cleave a specific phosphodiester bond on an mRNA molecule.[8] Lower eukaryotes and a few bacteria contain group I and group II ribozymes.[8] These motifs can self-splice by cleaving and joining together phosphodiester bonds.[8] The last ribozyme motif, the RNase P ribozyme, is found in Escherichia coli and is known for its ability to cleave the phosphodiester bonds of several tRNA precursors when joined to a protein cofactor.[8]
The general catalytic mechanism used by ribozymes is similar to the mechanism used by protein ribonucleases.[10] These catalytic RNA molecules bind to a specific site and attack the neighboring phosphate in the RNA backbone with their 2' oxygen, which acts as a nucleophile, resulting in the formation of cleaved products with a 2'3'-cyclic phosphate and a 5' hydroxyl terminal end.[10] This catalytic mechanism has been increasingly used by scientists to perform sequence-specific cleavage of target mRNA molecules. In addition, attempts are being made to use ribozymes to produce gene silencing therapeutics, which would silence genes that are responsible for causing diseases.[11]
RNA interference
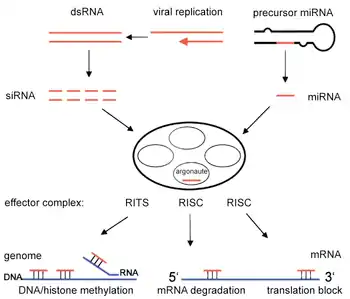
RNA interference (RNAi) is a natural process used by cells to regulate gene expression. It was discovered in 1998 by Andrew Fire and Craig Mello, who won the Nobel Prize for their discovery in 2006.[12] The process to silence genes first begins with the entrance of a double-stranded RNA (dsRNA) molecule into the cell, which triggers the RNAi pathway.[12] The double-stranded molecule is then cut into small double-stranded fragments by an enzyme called Dicer.[12] These small fragments, which include small interfering RNAs (siRNA) and microRNA (miRNA), are approximately 21–23 nucleotides in length.[12][13] The fragments integrate into a multi-subunit protein called the RNA-induced silencing complex, which contains Argonaute proteins that are essential components of the RNAi pathway.[12][13] One strand of the molecule, called the "guide" strand, binds to RISC, while the other strand, known as the "passenger" strand is degraded.[12][13] The guide or antisense strand of the fragment that remains bound to RISC directs the sequence-specific silencing of the target mRNA molecule.[13] The genes can be silenced by siRNA molecules that cause the endonucleatic cleavage of the target mRNA molecules or by miRNA molecules that suppress translation of the mRNA molecule.[13] With the cleavage or translational repression of the mRNA molecules, the genes that form them are rendered essentially inactive.[12] RNAi is thought to have evolved as a cellular defense mechanism against invaders, such as RNA viruses, or to combat the proliferation of transposons within a cell's DNA.[12] Both RNA viruses and transposons can exist as double-stranded RNA and lead to the activation of RNAi.[12] Currently, siRNAs are being widely used to suppress specific gene expression and to assess the function of genes. Companies utilizing this approach include Alnylam, Sanofi,[14] Arrowhead, Discerna,[15] and Persomics,[16] among others.
Three prime untranslated regions and microRNAs
Three prime untranslated regions (3'UTRs) of messenger RNAs (mRNAs) often contain regulatory sequences that post-transcriptionally cause gene silencing. Such 3'-UTRs often contain both binding sites for microRNAs (miRNAs) as well as for regulatory proteins. By binding to specific sites within the 3'-UTR, a large number of specific miRNAs decrease gene expression of their particular target mRNAs by either inhibiting translation or directly causing degradation of the transcript, using a mechanism similar to RNA interference (see MicroRNA). The 3'-UTR also may have silencer regions that bind repressor proteins that inhibit the expression of an mRNA.
The 3'-UTR often contains microRNA response elements (MREs). MREs are sequences to which miRNAs bind and cause gene silencing. These are prevalent motifs within 3'-UTRs. Among all regulatory motifs within the 3'-UTRs (e.g. including silencer regions), MREs make up about half of the motifs.
As of 2014, the miRBase web site,[17] an archive of miRNA sequences and annotations, listed 28,645 entries in 233 biologic species. Of these, 1,881 miRNAs were in annotated human miRNA loci. miRNAs were predicted to each have an average of about four hundred target mRNAs (causing gene silencing of several hundred genes).[18] Freidman et al.[18] estimate that >45,000 miRNA target sites within human mRNA 3'UTRs are conserved above background levels, and >60% of human protein-coding genes have been under selective pressure to maintain pairing to miRNAs.
Direct experiments show that a single miRNA can reduce the stability of hundreds of unique mRNAs.[19] Other experiments show that a single miRNA may repress the production of hundreds of proteins, but that this repression often is relatively mild (less than 2-fold).[20][21]
The effects of miRNA dysregulation of gene expression seem to be important in cancer.[22] For instance, in gastrointestinal cancers, nine miRNAs have been identified as epigenetically altered and effective in down regulating DNA repair enzymes.[23]
The effects of miRNA dysregulation of gene expression also seem to be important in neuropsychiatric disorders, such as schizophrenia, bipolar disorder, major depression, Parkinson's disease, Alzheimer's disease and autism spectrum disorders.[24][25][26]
Applications
Medical research
Gene silencing techniques have been widely used by researchers to study genes associated with disorders. These disorders include cancer, infectious diseases, respiratory diseases, and neurodegenerative disorders. Gene silencing is also currently being used in drug discovery efforts, such as synthetic lethality, high-throughput screening, and miniaturized RNAi screens.
Cancer
RNA interference has been used to silence genes associated with several cancers. In in vitro studies of chronic myelogenous leukemia (CML), siRNA was used to cleave the fusion protein, BCR-ABL, which prevents the drug Gleevec (imatinib) from binding to the cancer cells.[27] Cleaving the fusion protein reduced the amount of transformed hematopoietic cells that spread throughout the body by increasing the sensitivity of the cells to the drug.[27] RNA interference can also be used to target specific mutants. For instance, siRNAs were able to bind specifically to tumor suppressor p53 molecules containing a single point mutation and destroy it, while leaving the wild-type suppressor intact.[28]
Receptors involved in mitogenic pathways that lead to the increased production of cancer cells there have also been targeted by siRNA molecules. The chemokine receptor chemokine receptor 4 (CXCR4), associated with the proliferation of breast cancer, was cleaved by siRNA molecules that reduced the number of divisions commonly observed by the cancer cells.[29] Researchers have also used siRNAs to selectively regulate the expression of cancer-related genes. Antiapoptotic proteins, such as clusterin and survivin, are often expressed in cancer cells.[30][31] Clusterin and survivin-targeting siRNAs were used to reduce the number of antiapoptotic proteins and, thus, increase the sensitivity of the cancer cells to chemotherapy treatments.[30][31] In vivo studies are also being increasingly utilized to study the potential use of siRNA molecules in cancer therapeutics. For instance, mice implanted with colon adenocarcinoma cells were found to survive longer when the cells were pretreated with siRNAs that targeted B-catenin in the cancer cells.[32]
Viruses
Viral genes and host genes that are required for viruses to replicate or enter the cell, or that play an important role in the life cycle of the virus are often targeted by antiviral therapies. RNAi has been used to target genes in several viral diseases, such as the human immunodeficiency virus (HIV) and hepatitis.[33][34] In particular, siRNA was used to silence the primary HIV receptor chemokine receptor 5 (CCR5).[35] This prevented the virus from entering the human peripheral blood lymphocytes and the primary hematopoietic stem cells.[35][36] A similar technique was used to decrease the amount of the detectable virus in hepatitis B and C infected cells. In hepatitis B, siRNA silencing was used to target the surface antigen on the hepatitis B virus and led to a decrease in the number of viral components.[37] In addition, siRNA techniques used in hepatitis C were able to lower the amount of the virus in the cell by 98%.[38][39]
RNA interference has been in commercial use to control virus diseases of plants for over 20 years (see Plant disease resistance). In 1986–1990, multiple examples of "coat protein-mediated resistance" against plant viruses were published, before RNAi had been discovered.[40] In 1993, work with tobacco etch virus first demonstrated that host organisms can target specific virus or mRNA sequences for degradation, and that this activity is the mechanism behind some examples of virus resistance in transgenic plants.[41][42] The discovery of small interfering RNAs (the specificity determinant in RNA-mediated gene silencing) also utilized virus-induced post-transcriptional gene silencing in plants.[43] By 1994, transgenic squash varieties had been generated expressing coat protein genes from three different viruses, providing squash hybrids with field-validated multiviral resistance that remain in commercial use at present. Potato lines expressing viral replicase sequences that confer resistance to potato leafroll virus were sold under the trade names NewLeaf Y and NewLeaf Plus, and were widely accepted in commercial production in 1999–2001, until McDonald's Corp. decided not to purchase GM potatoes and Monsanto decided to close their NatureMark potato business.[44] Another frequently cited example of virus resistance mediated by gene silencing involves papaya, where the Hawaiian papaya industry was rescued by virus-resistant GM papayas produced and licensed by university researchers rather than a large corporation.[45] These papayas also remain in use at present, although not without significant public protest,[46][47] which is notably less evident in medical uses of gene silencing.
Gene silencing techniques have also been used to target other viruses, such as the human papilloma virus, the West Nile virus, and the Tulane virus. The E6 gene in tumor samples retrieved from patients with the human papilloma virus was targeted and found to cause apoptosis in the infected cells.[48] Plasmid siRNA expression vectors used to target the West Nile virus were also able to prevent the replication of viruses in cell lines.[49] In addition, siRNA has been found to be successful in preventing the replication of the Tulane virus, part of the virus family Caliciviridae, by targeting both its structural and non-structural genes.[50] By targeting the NTPase gene, one dose of siRNA 4 hours pre-infection was shown to control Tulane virus replication for 48 hours post-infection, reducing the viral titer by up to 2.6 logarithms.[50] Although the Tulane virus is species-specific and does not affect humans, it has been shown to be closely related to the human norovirus, which is the most common cause of acute gastroenteritis and food-borne disease outbreaks in the United States.[51] Human noroviruses are notorious for being difficult to study in the laboratory, but the Tulane virus offers a model through which to study this family of viruses for the clinical goal of developing therapies that can be used to treat illnesses caused by human norovirus.
Bacteria
Unlike viruses, bacteria are not as susceptible to silencing by siRNA.[52] This is largely due to how bacteria replicate. Bacteria replicate outside of the host cell and do not contain the necessary machinery for RNAi to function.[52] However, bacterial infections can still be suppressed by siRNA by targeting the host genes that are involved in the immune response caused by the infection or by targeting the host genes involved in mediating the entry of bacteria into cells.[52][53] For instance, siRNA was used to reduce the amount of pro-inflammatory cytokines expressed in the cells of mice treated with lipopolysaccharide (LPS).[52][54] The reduced expression of the inflammatory cytokine, tumor necrosis factor α (TNFα), in turn, caused a reduction in the septic shock felt by the LPS-treated mice.[54] In addition, siRNA was used to prevent the bacteria, Psueomonas aeruginosa, from invading murine lung epithelial cells by knocking down the caveolin-2 (CAV2) gene.[55] Thus, though bacteria cannot be directly targeted by siRNA mechanisms, they can still be affected by siRNA when the components involved in the bacterial infection are targeted.
Respiratory diseases
Ribozymes, antisense oligonucleotides, and more recently RNAi have been used to target mRNA molecules involved in asthma.[53][56] These experiments have suggested that siRNA may be used to combat other respiratory diseases, such as chronic obstructive pulmonary disease (COPD) and cystic fibrosis.[53] COPD is characterized by goblet cell hyperplasia and mucus hypersecretion.[57] Mucus secretion was found to be reduced when the transforming growth factor (TGF)-α was targeted by siRNA in NCI-H292 human airway epithelial cells.[58] In addition to mucus hypersecretion, chronic inflammation and damaged lung tissue are characteristic of COPD and asthma. The transforming growth factor TGF-β is thought to play a role in these manifestations.[59][60] As a result, when interferon (IFN)-γ was used to knock down TGF-β, fibrosis of the lungs, caused by damage and scarring to lung tissue, was improved.[61][62]
Huntington's disease
Huntington's disease (HD) results from a mutation in the huntingtin gene that causes an excess of CAG repeats.[63] The gene then forms a mutated huntingtin protein with polyglutamine repeats near the amino terminus.[64] This disease is incurable and known to cause motor, cognitive, and behavioral deficits.[65] Researchers have been looking to gene silencing as a potential therapeutic for HD.
Gene silencing can be used to treat HD by targeting the mutant huntingtin protein. The mutant huntingtin protein has been targeted through gene silencing that is allele specific using allele specific oligonucleotides. In this method, the antisense oligonucleotides are used to target single nucleotide polymorphism (SNPs), which are single nucleotide changes in the DNA sequence, since HD patients have been found to share common SNPs that are associated with the mutated huntingtin allele. It has been found that approximately 85% of patients with HD can be covered when three SNPs are targeted. In addition, when antisense oligonucleotides were used to target an HD-associated SNP in mice, there was a 50% decrease in the mutant huntingtin protein.[63]
Non-allele specific gene silencing using siRNA molecules has also been used to silence the mutant huntingtin proteins. Through this approach, instead of targeting SNPs on the mutated protein, all of the normal and mutated huntingtin proteins are targeted. When studied in mice, it was found that siRNA could reduce the normal and mutant huntingtin levels by 75%. At this level, they found that the mice developed improved motor control and a longer survival rate when compared to the controls.[63] Thus, gene silencing methods may prove to be beneficial in treating HD.
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS), also called Lou Gehrig's disease, is a motor neuron disease that affects the brain and spinal cord. The disease causes motor neurons to degenerate, which eventually leads to neuron death and muscular degeneration.[66] Hundreds of mutations in the Cu/Zn superoxide dismutase (SOD1) gene have been found to cause ALS.[67] Gene silencing has been used to knock down the SOD1 mutant that is characteristic of ALS.[67][68] In specific, siRNA molecules have been successfully used to target the SOD1 mutant gene and reduce its expression through allele-specific gene silencing.[67][69]
Therapeutics challenges

There are several challenges associated with gene silencing therapies, including delivery and specificity for targeted cells. For instance, for treatment of neurodegenerative disorders, molecules for a prospective gene silencing therapy must be delivered to the brain. The blood-brain barrier makes it difficult to deliver molecules into the brain through the bloodstream by preventing the passage of the majority of molecules that are injected or absorbed into the blood.[63][64] Thus, researchers have found that they must directly inject the molecules or implant pumps that push them into the brain.[63]
Once inside the brain, however, the molecules must move inside of the targeted cells. In order to efficiently deliver siRNA molecules into the cells, viral vectors can be used.[63][65] Nevertheless, this method of delivery can also be problematic as it can elicit an immune response against the molecules. In addition to delivery, specificity has also been found to be an issue in gene silencing. Both antisense oligonucleotides and siRNA molecules can potentially bind to the wrong mRNA molecule.[63] Thus, researchers are searching for more efficient methods to deliver and develop specific gene silencing therapeutics that are still safe and effective.
References
- Redberry, Grace (2006). Gene silencing : new research. New York: Nova Science Publishers. ISBN 9781594548321.
- "Gene Silencing". National Center for Biotechnology Information. Retrieved 11 November 2013.
- Hood E (March 2004). "RNAi: What's all the noise about gene silencing?". Environmental Health Perspectives. 112 (4): A224–9. doi:10.1289/ehp.112-a224. PMC 1241909. PMID 15033605.
- Mocellin S, Provenzano M (November 2004). "RNA interference: learning gene knock-down from cell physiology". Journal of Translational Medicine. 2 (1): 39. doi:10.1186/1479-5876-2-39. PMC 534783. PMID 15555080.
- Kole R, Krainer AR, Altman S (February 2012). "RNA therapeutics: beyond RNA interference and antisense oligonucleotides". Nature Reviews. Drug Discovery. 11 (2): 125–40. doi:10.1038/nrd3625. PMC 4743652. PMID 22262036.
- Dias N, Stein CA (March 2002). "Antisense oligonucleotides: basic concepts and mechanisms". Molecular Cancer Therapeutics. 1 (5): 347–55. PMID 12489851.
- Kurreck J (March 2004). "Antisense and RNA interference approaches to target validation in pain research". Current Opinion in Drug Discovery & Development. 7 (2): 179–87. PMID 15603251.
- Phylactou, L. (1 September 1998). "Ribozymes as therapeutic tools for genetic disease". Human Molecular Genetics. 7 (10): 1649–1653. doi:10.1093/hmg/7.10.1649. PMID 9735387.
- Shampo MA, Kyle RA, Steensma DP (October 2012). "Sidney Altman--Nobel laureate for work with RNA". Mayo Clinic Proceedings. 87 (10): e73. doi:10.1016/j.mayocp.2012.01.022. PMC 3498233. PMID 23036683.
- Doherty EA, Doudna JA (1 June 2001). "Ribozyme structures and mechanisms". Annual Review of Biophysics and Biomolecular Structure. 30 (1): 457–75. doi:10.1146/annurev.biophys.30.1.457. PMID 11441810.
- Tollefsbol, edited by Trygve O. (2007). Biological aging methods and protocols. Totowa, N.J.: Humana Press. ISBN 9781597453615.CS1 maint: extra text: authors list (link)
- "RNA Interference Fact Sheet". National Institutes of Health. Archived from the original on November 25, 2013. Retrieved 24 November 2013.
- Wilson RC, Doudna JA (2013). "Molecular mechanisms of RNA interference". Annual Review of Biophysics. 42: 217–39. doi:10.1146/annurev-biophys-083012-130404. PMC 5895182. PMID 23654304.
- "Big Pharma's Turn On RNAi Shows That New Technologies Don't Guarantee R&D Success". Forbes. Retrieved 2015-10-11.
- "The Second Coming of RNAi | The Scientist Magazine®". The Scientist. Retrieved 2015-10-11.
- "Products | Persomics". www.persomics.com. Retrieved 2015-10-11.
- miRBase.org
- Friedman RC, Farh KK, Burge CB, Bartel DP (January 2009). "Most mammalian mRNAs are conserved targets of microRNAs". Genome Research. 19 (1): 92–105. doi:10.1101/gr.082701.108. PMC 2612969. PMID 18955434.
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (February 2005). "Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs". Nature. 433 (7027): 769–73. Bibcode:2005Natur.433..769L. doi:10.1038/nature03315. PMID 15685193. S2CID 4430576.
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (September 2008). "Widespread changes in protein synthesis induced by microRNAs". Nature. 455 (7209): 58–63. Bibcode:2008Natur.455...58S. doi:10.1038/nature07228. PMID 18668040. S2CID 4429008.
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP (September 2008). "The impact of microRNAs on protein output". Nature. 455 (7209): 64–71. Bibcode:2008Natur.455...64B. doi:10.1038/nature07242. PMC 2745094. PMID 18668037.
- Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, Marques MM (July 2011). "Mechanisms and role of microRNA deregulation in cancer onset and progression". Genetics and Molecular Biology. 34 (3): 363–70. doi:10.1590/S1415-47572011000300001. PMC 3168173. PMID 21931505.
- Bernstein C, Bernstein H (May 2015). "Epigenetic reduction of DNA repair in progression to gastrointestinal cancer". World Journal of Gastrointestinal Oncology. 7 (5): 30–46. doi:10.4251/wjgo.v7.i5.30. PMC 4434036. PMID 25987950.
- Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L (2014). "Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders". Frontiers in Cellular Neuroscience. 8: 75. doi:10.3389/fncel.2014.00075. PMC 3949217. PMID 24653674.
- Mellios N, Sur M (2012). "The Emerging Role of microRNAs in Schizophrenia and Autism Spectrum Disorders". Frontiers in Psychiatry. 3: 39. doi:10.3389/fpsyt.2012.00039. PMC 3336189. PMID 22539927.
- Geaghan M, Cairns MJ (August 2015). "MicroRNA and Posttranscriptional Dysregulation in Psychiatry". Biological Psychiatry. 78 (4): 231–9. doi:10.1016/j.biopsych.2014.12.009. PMID 25636176. S2CID 5730697.
- Chen J, Wall NR, Kocher K, Duclos N, Fabbro D, Neuberg D, Griffin JD, Shi Y, Gilliland DG (June 2004). "Stable expression of small interfering RNA sensitizes TEL-PDGFbetaR to inhibition with imatinib or rapamycin". The Journal of Clinical Investigation. 113 (12): 1784–91. doi:10.1172/JCI20673. PMC 420507. PMID 15199413.
- Martinez LA, Naguibneva I, Lehrmann H, Vervisch A, Tchénio T, Lozano G, Harel-Bellan A (November 2002). "Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways". Proceedings of the National Academy of Sciences of the United States of America. 99 (23): 14849–54. Bibcode:2002PNAS...9914849M. doi:10.1073/pnas.222406899. PMC 137507. PMID 12403821.
- Lapteva N, Yang AG, Sanders DE, Strube RW, Chen SY (January 2005). "CXCR4 knockdown by small interfering RNA abrogates breast tumor growth in vivo". Cancer Gene Therapy. 12 (1): 84–9. doi:10.1038/sj.cgt.7700770. PMID 15472715. S2CID 23402257.
- July LV, Beraldi E, So A, Fazli L, Evans K, English JC, Gleave ME (March 2004). "Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo". Molecular Cancer Therapeutics. 3 (3): 223–32. PMID 15026542.
- Ning S, Fuessel S, Kotzsch M, Kraemer K, Kappler M, Schmidt U, Taubert H, Wirth MP, Meye A (October 2004). "siRNA-mediated down-regulation of survivin inhibits bladder cancer cell growth". International Journal of Oncology. 25 (4): 1065–71. doi:10.3892/ijo.25.4.1065 (inactive 2021-01-10). PMID 15375557.CS1 maint: DOI inactive as of January 2021 (link)
- Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB (April 2003). "Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells". Clinical Cancer Research. 9 (4): 1291–300. PMID 12684397.
- Dave RS, Pomerantz RJ (December 2004). "Antiviral effects of human immunodeficiency virus type 1-specific small interfering RNAs against targets conserved in select neurotropic viral strains". Journal of Virology. 78 (24): 13687–96. doi:10.1128/JVI.78.24.13687-13696.2004. PMC 533941. PMID 15564478.
- Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD (March 2003). "RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells". Proceedings of the National Academy of Sciences of the United States of America. 100 (5): 2783–8. Bibcode:2003PNAS..100.2783W. doi:10.1073/pnas.252758799. PMC 151418. PMID 12594341.
- Qin XF, An DS, Chen IS, Baltimore D (January 2003). "Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5". Proceedings of the National Academy of Sciences of the United States of America. 100 (1): 183–8. Bibcode:2002PNAS..100..183Q. doi:10.1073/pnas.232688199. PMC 140921. PMID 12518064.
- Li MJ, Bauer G, Michienzi A, Yee JK, Lee NS, Kim J, Li S, Castanotto D, Zaia J, Rossi JJ (August 2003). "Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs". Molecular Therapy. 8 (2): 196–206. doi:10.1016/s1525-0016(03)00165-5. PMID 12907142.
- Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E (November 2003). "Small interfering RNA inhibits hepatitis B virus replication in mice". Molecular Therapy. 8 (5): 769–76. doi:10.1016/s1525-0016(03)00244-2. PMID 14599810.
- Randall G, Grakoui A, Rice CM (January 2003). "Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs". Proceedings of the National Academy of Sciences of the United States of America. 100 (1): 235–40. Bibcode:2002PNAS..100..235R. doi:10.1073/pnas.0235524100. PMC 140937. PMID 12518066.
- Randall G, Rice CM (June 2004). "Interfering with hepatitis C virus RNA replication". Virus Research. 102 (1): 19–25. doi:10.1016/j.virusres.2004.01.011. PMID 15068876.
- Beachy RN, Loesch-Fries S, Tumer NE (1990). "Coat Protein-Mediated Resistance Against Virus Infection". Annual Review of Phytopathology. 28: 451–472. doi:10.1146/annurev.py.28.090190.002315.
- Lindbo JA, Dougherty WG (2005). "Plant pathology and RNAi: a brief history". Annual Review of Phytopathology. 43: 191–204. doi:10.1146/annurev.phyto.43.040204.140228. PMID 16078882.
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (December 1993). "Induction of a Highly Specific Antiviral State in Transgenic Plants: Implications for Regulation of Gene Expression and Virus Resistance". The Plant Cell. 5 (12): 1749–1759. doi:10.1105/tpc.5.12.1749. PMC 160401. PMID 12271055.
- Hamilton AJ, Baulcombe DC (October 1999). "A species of small antisense RNA in posttranscriptional gene silencing in plants". Science. 286 (5441): 950–2. doi:10.1126/science.286.5441.950. PMID 10542148.
- Kaniewski WK, Thomas PE (2004). "The Potato Story". AgBioForum. 7 (1&2): 41–46.
- Ferreira, S. A.; Pitz, K. Y.; Manshardt, R.; Zee, F.; Fitch, M.; Gonsalves, D. (2002). "Virus Coat Protein Transgenic Papaya Provides Practical Control ofPapaya ringspot virusin Hawaii". Plant Disease. 86 (2): 101–105. doi:10.1094/PDIS.2002.86.2.101. PMID 30823304.
- "Papaya: A GMO success story". Archived from the original on 2015-06-10. Retrieved 2016-08-30.
- "Papaya in the Crosshairs: A Heated Island Battle Over GMOs - Modern Farmer". 19 December 2013.
- Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F (September 2003). "siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells". Oncogene. 22 (38): 5938–45. doi:10.1038/sj.onc.1206894. PMID 12955072. S2CID 21504155.
- McCown M, Diamond MS, Pekosz A (September 2003). "The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative- and positive-strand RNA viruses". Virology. 313 (2): 514–24. doi:10.1016/s0042-6822(03)00341-6. PMID 12954218.
- Fan Q, Wei C, Xia M, Jiang X (January 2013). "Inhibition of Tulane virus replication in vitro with RNA interference". Journal of Medical Virology. 85 (1): 179–86. doi:10.1002/jmv.23340. PMC 3508507. PMID 23154881.
- "Norovirus Overview". Center for Disease Control and Prevention. 2018-12-21.
- Lieberman J, Song E, Lee SK, Shankar P (September 2003). "Interfering with disease: opportunities and roadblocks to harnessing RNA interference". Trends in Molecular Medicine. 9 (9): 397–403. doi:10.1016/s1471-4914(03)00143-6. PMC 7128953. PMID 13129706.
- Leung RK, Whittaker PA (August 2005). "RNA interference: from gene silencing to gene-specific therapeutics". Pharmacology & Therapeutics. 107 (2): 222–39. doi:10.1016/j.pharmthera.2005.03.004. PMC 7112686. PMID 15908010.
- Sørensen DR, Sioud M (2010). "Systemic delivery of synthetic siRNAs". RNA Therapeutics. Methods in Molecular Biology. 629. pp. 87–91. doi:10.1007/978-1-60761-657-3_6. ISBN 978-1-60761-656-6. PMID 20387144.
- Zaas DW, Duncan MJ, Li G, Wright JR, Abraham SN (February 2005). "Pseudomonas invasion of type I pneumocytes is dependent on the expression and phosphorylation of caveolin-2". The Journal of Biological Chemistry. 280 (6): 4864–72. doi:10.1074/jbc.M411702200. PMID 15545264. S2CID 43122091.
- Popescu FD, Popescu F (September 2007). "A review of antisense therapeutic interventions for molecular biological targets in asthma". Biologics. 1 (3): 271–83. PMC 2721314. PMID 19707336.
- Pistelli R, Lange P, Miller DL (May 2003). "Determinants of prognosis of COPD in the elderly: mucus hypersecretion, infections, cardiovascular comorbidity". The European Respiratory Journal. Supplement. 40: 10s–14s. doi:10.1183/09031936.03.00403403. PMID 12762568. S2CID 19006320.
- Shao MX, Nakanaga T, Nadel JA (August 2004). "Cigarette smoke induces MUC5AC mucin overproduction via tumor necrosis factor-alpha-converting enzyme in human airway epithelial (NCI-H292) cells". American Journal of Physiology. Lung Cellular and Molecular Physiology. 287 (2): L420–7. doi:10.1152/ajplung.00019.2004. PMID 15121636.
- Rennard SI (November 1999). "Inflammation and repair processes in chronic obstructive pulmonary disease". American Journal of Respiratory and Critical Care Medicine. 160 (5 Pt 2): S12–6. doi:10.1164/ajrccm.160.supplement_1.5. PMID 10556162.
- Sacco O, Silvestri M, Sabatini F, Sale R, Defilippi AC, Rossi GA (2004). "Epithelial cells and fibroblasts: structural repair and remodelling in the airways". Paediatric Respiratory Reviews. 5 Suppl A: S35–40. doi:10.1016/s1526-0542(04)90008-5. PMID 14980241.
- "Pulmonary Fibrosis". Mayo Clinic. Retrieved 13 December 2013.
- Gurujeyalakshmi G, Giri SN (Sep–Oct 1995). "Molecular mechanisms of antifibrotic effect of interferon gamma in bleomycin-mouse model of lung fibrosis: downregulation of TGF-beta and procollagen I and III gene expression". Experimental Lung Research. 21 (5): 791–808. doi:10.3109/01902149509050842. PMID 8556994.
- "Gene Silencing". HOPES - Huntington's Outreach Project for Education, at Stanford. Stanford University. 2012-04-05. Retrieved 13 December 2013.
- Mantha N, Das SK, Das NG (September 2012). "RNAi-based therapies for Huntington's disease: delivery challenges and opportunities". Therapeutic Delivery. 3 (9): 1061–76. doi:10.4155/tde.12.80. PMID 23035592.
- Harper SQ (August 2009). "Progress and challenges in RNA interference therapy for Huntington disease". Archives of Neurology. 66 (8): 933–8. doi:10.1001/archneurol.2009.180. PMID 19667213.
- "What is ALS?". The ALS Association.
- Geng CM, Ding HL (February 2008). "Double-mismatched siRNAs enhance selective gene silencing of a mutant ALS-causing allele". Acta Pharmacologica Sinica. 29 (2): 211–6. doi:10.1111/j.1745-7254.2008.00740.x. PMID 18215350. S2CID 24809180.
- Boulis, Nicholas. "Gene Therapy for Motor Neuron Disease". Society for Neuroscience. Retrieved 13 December 2013.
- Ding H, Schwarz DS, Keene A, Affar el B, Fenton L, Xia X, Shi Y, Zamore PD, Xu Z (August 2003). "Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis". Aging Cell. 2 (4): 209–17. doi:10.1046/j.1474-9728.2003.00054.x. PMID 12934714. S2CID 31752201.
- Petition for Determination of Nonregulated Status: Arctic™ Apple (Malus x domestica) Events GD743 and GS784. United States Department of Agriculture – Animal and Plant Health Inspection Service. Retrieved 2012-08-03.
- "Apple-to-apple transformation". Okanagan Specialty Fruits. Archived from the original on 2013-09-25. Retrieved 2012-08-03.
External links
- RNAiAtlas - database of siRNA libraries and their target analysis resultsArchived February 10, 2015, at the Wayback Machine.
- Science project: Transgenic apple varieties Approaches to preventing outcrossing – possible effects on micro-organisms
- Gene+silencing at the US National Library of Medicine Medical Subject Headings (MeSH)
- Research project: New Cost-effective method for gene silencing
- van Leeuwen F, Gottschling DE (2002). "Assays for gene silencing in yeast". Guide to Yeast Genetics and Molecular and Cell Biology - Part B. Methods in Enzymology. 350. pp. 165–86. doi:10.1016/S0076-6879(02)50962-9. ISBN 9780121822538. PMID 12073311.