HLA-A66
HLA-A66 (A66) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α66 subset of HLA-A α-chains. For A66, the alpha "A" chain are encoded by the HLA-A*66 allele group and the β-chain are encoded by B2M locus.[1] A66 and A*66 are almost synonymous in meaning. A66 is a split antigen of the broad antigen serotype A10. A66 is a sister serotype of A25, A26, A34, and A43.
| HLA-A66 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||||||||
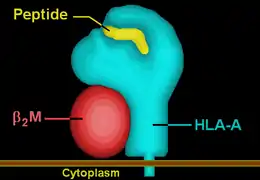 HLA-A66 | ||||||||||||||||
| About | ||||||||||||||||
| Protein | transmembrane receptor/ligand | |||||||||||||||
| Structure | αβ heterodimer | |||||||||||||||
| Subunits | HLA-A*66--, β2-microglobulin | |||||||||||||||
| Older names | A10 | |||||||||||||||
| Subtypes | ||||||||||||||||
| ||||||||||||||||
| Rare alleles | ||||||||||||||||
| ||||||||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||||||||
A66 is more common in Africa and Southwest Europe. A66 (A*6601) is believed to have been formed by a single gene conversion between another HLA-A and the A*2601 allele.[2].
Serotype
| A*66 | A66 | A10 | A26 | A34 | Sample |
| allele | % | % | % | % | size (N) |
| *6601 | 20 | 10 | 61 | 8 | 799 |
| *6602 | 38 | 10 | 4 | 27 | 205 |
| *6603 | 32 | 7 | 7 | 18 | 28 |
A66 serotyping is poor. A*6601 is also sometimes recognized by A25, and A*6602 is often recognized by A74.
A*6601 Frequencies
| freq | ||
| ref. | Population | (%) |
| [4] | Cameroon Sawa | 7.7 |
| [4] | Kenya Luo | 6.8 |
| [4] | Cameroon Sawa | 7.7 |
| [4] | Cameroon Bakola Pygmy | 5.8 |
| [4] | Cameroon Baka Pygmy | 5.0 |
| [4] | Kenya Nandi | 5.0 |
| [4] | Zimbabwe Harare Shona | 0.2 |
| [4] | Cameroon Baka Pygmy | 5.0 |
| [4] | Cameroon Beti | 4.6 |
| [4] | Cameroon Bamileke | 4.5 |
| [4] | India West Bhils | 4.0 |
| [4] | Zimbabwe Harere Shona | 3.8 |
| [4] | Uganda Kampala | 3.8 |
| [4] | India Mumbai Marathas | 2.5 |
| [4] | Cape Verde Northwestern | 2.5 |
| [4] | Czech republic | 2.4 |
| [4] | Morocco Nador Metalsa | 2.1 |
| [4] | Central Portugal | 2.0 |
| [4] | India West Parsis | 2.0 |
| [4] | South African Natal Zulu | 2.0 |
| [4] | Kenya | 1.7 |
| [4] | Tunisia Tunis | 1.7 |
| [4] | Guinea Bissau | 1.5 |
| [4] | Georgia Tibilisi | 1.4 |
| [4] | Zambia Lusaka | 1.2 |
| [4] | Italy Bergamo | 1.1 |
| [4] | Pakistan Karachi Parsi | 1.1 |
| [4] | Oman | 0.8 |
| [4] | Sudanese | 0.8 |
| [4] | Belgium | 0.5 |
| [4] | Southeast France | 0.4 |
| [4] | Mongolia Buriat | 0.4 |
| [4] | Wales | 0.2 |
| freq | ||
| ref. | Population | (%) |
| [4] | South African Natal Zulu | 1.5 |
| [4] | Cameroon Yaounde | 1.1 |
| [4] | Cameroon Bamileke | 0.6 |
| [4] | Senegal Niokholo Mandenka | 0.5 |
| [4] | Kenya Nandi | 0.4 |
| [4] | Zimbabwe Harare Shona | 0.2 |
| [4] | Wales | 0.03 |
| HLA A*6603 frequencies | ||
| freq | ||
| ref. | Population | (%) |
| [4] | Cameroon Pygmy Baka | 10.0 |
| [4] | Cameroon Bakola Pygmy | 9.0 |
| [4] | Cameroon Sawa | 7.7 |
| [4] | CAR Mbenzele Pygmy | 2.8 |
| [4] | Zimbabwe Harare Shona | 0.2 |
References
- Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin". Tissue Antigens. 11 (2): 96–112. doi:10.1111/j.1399-0039.1978.tb01233.x. PMID 77067.
- Madrigal JA, Hildebrand WH, Belich MP, et al. (1993). "Structural diversity in the HLA-A10 family of alleles: correlations with serology". Tissue Antigens. 41 (2): 72–80. doi:10.1111/j.1399-0039.1993.tb01982.x. PMID 8475492.
- Allele Query Form IMGT/HLA - European Bioinformatics Institute
- Middleton D, Menchaca L, Rood H, Komerofsky R (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–7. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660. External link in
|title=(help)
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.