HLA-A80
HLA-A80 (A80) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α80 subset of HLA-A α-chains. For A80, the alpha "A" chain are encoded by the HLA-A*80 allele group and the β-chain are encoded by B2M locus.[1] This group currently is dominated by A*8001. A80 and A*80 are almost synonymous in meaning.
| HLA-A80 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||||||||
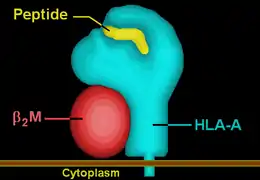 HLA-A80 | ||||||||||||||||
| About | ||||||||||||||||
| Protein | transmembrane receptor/ligand | |||||||||||||||
| Structure | αβ heterodimer | |||||||||||||||
| Subunits | HLA-A*8001, β2-microglobulin | |||||||||||||||
| Older names | AX"BG" | |||||||||||||||
| Subtypes | ||||||||||||||||
| ||||||||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||||||||
A80 is more common in West Central Africa. A80 is the least common HLA-A allele group.
Serotype
| A*80 | A80 | A10 | Sample |
| allele | % | % | size (N) |
| *8001 | 36 | 10 | 505 |
A80 HLA-A80 typing is poor. Most typing is done with SSP-PCR. Most of 8001 are detected with "Blank" serotype..
Distribution
| Study population | Freq. (in %)[3] |
|---|---|
| Senegal Niokholo Mandenka | 3.8 |
| Zambia Lusaka | 2.3 |
| Cameroon Bamileke | 1.9 |
| Mali Bandiagara | 1.5 |
| South Africa Tswana | 1.2 |
| Spain Menorca | 1.1 |
| Cameroon Bakola Pygmy | 1.0 |
| Burkina Faso Fulani | 1.0 |
| Trinidad Africans | 1.0 |
| Tunisia | 1.0 |
| Bulgaria | 0.9 |
| Cape Verde Southeastern I… | 0.8 |
| Guinea Bissau | 0.8 |
| USA African Americans | 0.8 |
| Peru Arequipa | 0.7 |
| Turkey class I | 0.7 |
| Cameroon Beti | 0.6 |
| Spain Catalonia Girona | 0.6 |
| USA Caucasians (3) | 0.6 |
| Brazil | 0.5 |
| Brazil Belo Horizonte | 0.5 |
| Brazil Pernambuco State | 0.5 |
| Croatia pop2 | 0.5 |
| Guatemala Mayans | 0.4 |
| Mongolia Buriat | 0.4 |
| Allele frequencies presented, only | |
Further reading
- Torimiro JN, Carr JK, Wolfe ND, et al. (2006). "HLA class I diversity among rural rainforest inhabitants in Cameroon: identification of A*2612-B*4407 haplotype". Tissue Antigens. 67 (1): 30–7. doi:10.1111/j.1399-0039.2005.00527.x. PMID 16451198.
- Sanchez-Mazas A, Steiner QG, Grundschober C, Tiercy JM (October 2000). "The molecular determination of HLA-Cw alleles in the Mandenka (West Africa) reveals a close genetic relationship between Africans and Europeans". Tissue Antigens. 56 (4): 303–12. doi:10.1034/j.1399-0039.2000.560402.x. PMID 11098930.
- Cao K, Moormann AM, Lyke KE, et al. (April 2004). "Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci". Tissue Antigens. 63 (4): 293–325. doi:10.1111/j.0001-2815.2004.00192.x. PMID 15009803.
- Torimiro JN, Carr JK, Wolfe ND, et al. (January 2006). "HLA class I diversity among rural rainforest inhabitants in Cameroon: identification of A*2612-B*4407 haplotype". Tissue Antigens. 67 (1): 30–7. doi:10.1111/j.1399-0039.2005.00527.x. PMID 16451198.
- Crespí C, Milà J, Martínez-Pomar N, et al. (October 2002). "HLA polymorphism in a Majorcan population of Jewish descent: comparison with Majorca, Minorca, Ibiza (Balearic Islands) and other Jewish communities". Tissue Antigens. 60 (4): 282–91. doi:10.1034/j.1399-0039.2002.600402.x. PMID 12472657.
References
- Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin". Tissue Antigens. 11 (2): 96–112. doi:10.1111/j.1399-0039.1978.tb01233.x. PMID 77067.
- Allele Query Form IMGT/HLA - European Bioinformatics Institute
- Middleton, D.; Menchaca, L.; Rood, H.; Komerofsky, R. (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–407. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.