Holobiont
A holobiont is an assemblage of a host and the many other species living in or around it, which together form a discrete ecological unit.[1] The components of a holobiont are individual species or bionts, while the combined genome of all bionts is the hologenome. The concept of the holobiont was initially defined by Dr. Lynn Margulis in her 1991 book Symbiosis as a Source of Evolutionary Innovation,[1] though the concept has subsequently evolved since the original definition.[2] Holobionts include the host, virome, microbiome, and other members, all of which contribute in some way to the function of the whole.[3][4] Well-studied holobionts include reef-building corals and humans.[5][6]

There is controversy over whether holobionts can be viewed as single evolutionary units.
Overview
A holobiont is a collection of species that are closely associated and have complex interactions, such as a plant species and the members of its microbiome.[1][7] Each species present in a holobiont is a biont, and the genomes of all bionts taken together are the hologenome, or the "comprehensive gene system" of the holobiont.[8] A holobiont typically includes a eukaryote host and all of the symbiotic viruses, bacteria, fungi, etc. that live on or inside it.[7]
Holobionts are distinct from superorganisms; superorganisms consist of many individuals, sometimes of the same species, and the term is commonly applied to eusocial insects.[9][10] An ant colony can be described as a superorganism, whereas an individual ant and its associated bacteria, fungi, etc. are a holobiont.[8] There is no doubt that symbiotic microorganisms are pivotal for the biology and ecology of the host by providing vitamins, energy and inorganic or organic nutrients, participating in defense mechanisms, or by driving the evolution of the host.[11][12] There is still some controversy surrounding these terms, and they have been used interchangeably in some publications.[6]
Holobiont components
Host: The host member of a holobiont is typically a multicellular eukaryote, such as a plant or human.[8] Notable hosts that are well-studied include humans,[13]</ref> corals,[5] and pine trees.[14]
Microbiome: The microbiome includes bacteria,[3] archaea,[15] microscopic fungi,[7] and microscopic protists.[3]
Virome: All of the viruses included in a holobiont are collectively referred to as the virome[16]
Fungi: Multicellular fungi can be included in holobionts, such as arbuscular mycorrhizal fungi (AMF) in the roots of plants.[7][4]
The holobiont phenotype
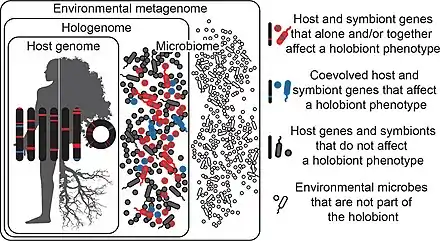
Holobionts are entities composed of a host and all of its symbiotic microbes.[2]
In the diagram, the symbiotic microbes that affect a holobiont's phenotype and have coevolved with the host are coloured blue, while those which affect the holobiont’s phenotype but have not coevolved with the host are coloured red. Those that do not affect the holobiont’s phenotype at all are coloured gray. Microbes may be transmitted vertically or horizontally, may be acquired from the environment, and can be constant or inconstant in the host.[2]
It follows that holobiont phenotypes can change in time and space as microbes come into and out of the holobiont. Microbes in the environment are not part of the holobiont (white). Hologenomes then encompass the genomes of the host and all of its microbes at any given time point, with individual genomes and genes falling into the same three functional categories of blue, red, and gray. Holobionts and hologenomes are entities, whereas coevolution or the evolution of host-symbiont interactions are processes.[2]
Example holobionts
Humans and wasps
[[File:Neo-Lamarckian inheritance of hologenome.svg|thumb|upright=2|left| {{center|Human holobiont[17]]]
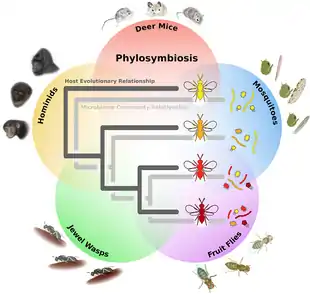
Plants
Although most work on host-microbe interactions has been focused on animal systems such as corals, sponges, or humans, there is a substantial body of literature on plant holobionts.[19] Plant-associated microbial communities impact both key components of the fitness of plants, growth and survival,[4] and are shaped by nutrient availability and plant defense mechanisms.[7] Several habitats have been described to harbor plant-associated microbes, including the rhizoplane (surface of root tissue), the rhizosphere (periphery of the roots), the endosphere (inside plant tissue), and the phyllosphere (total above-ground surface area).[12] The holobiont concept originally suggested that A significant fraction of the microbiome genome together with the host genome is transmitted from one generation to the next and thus can propagate unique properties of the holobiont".[20] In this regard, studies have shown that seeds can play such a role. Evidence of this process have been recently proven showing that the majority, up to 95%, of the seed microbiome is mistranslated across generations.[21]
The plant holobiont is relatively well-studied, with particular focus on agricultural species such as legumes and grains. Bacteria, fungi, archaea, protists, and viruses are all members of the plant holobiont.[3]
The bacteria phyla known to be part of the plant holobiont are Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria.[3] For example, nitrogen-fixers such as Azotobacter (Proteobacteria) and Bacillus (Firmicutes) greatly improve plant performance.[3]
Fungi of the phyla Ascomycota, Basidiomycota, and Glomeromycota colonize plant tissues and provide a variety of functions for the plant host.[3] Arbuscular mycorrhizal fungi (Glomeromycota), for instance, are common across plant groups and provide improved nutrient acquisition, temperature and drought resistance, and reduced pathogen load.[22] Epichloë species (Ascomycota) are part of the meadow fescue holobiont and provide herbivore resistance by producing ergot alkaloids, which cause ergotism in mammals.[23]
Protist members of the plant holobiont are less well-studied, with most knowledge oriented towards pathogens. However, there are examples of commensalistic plant-protist associations, such as Phytomonas (Trypanosomatidae).[24]
Marine
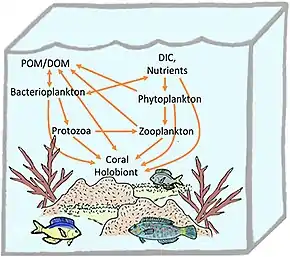
Reef-building corals are holobionts that include the coral itself (a eukaryotic invertebrate within class Anthozoa), photosynthetic dinoflagellates called zooxanthellae (Symbiodinium), and associated bacteria and viruses.[5] Co-evolutionary patterns exist for coral microbial communities and coral phylogeny.[25]
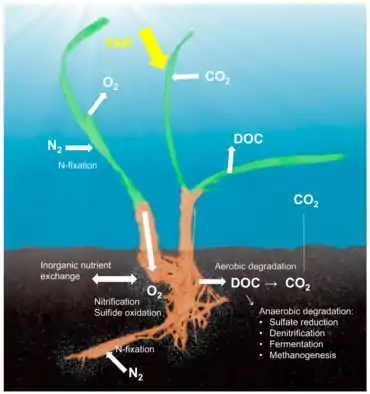
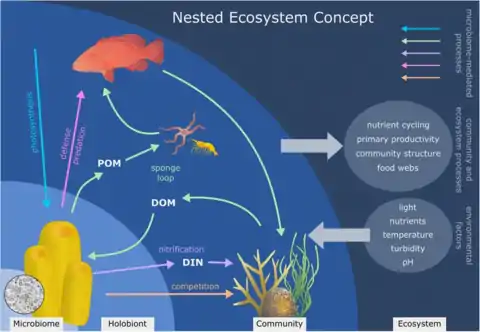 Sponge holobiont [27]
Sponge holobiont [27]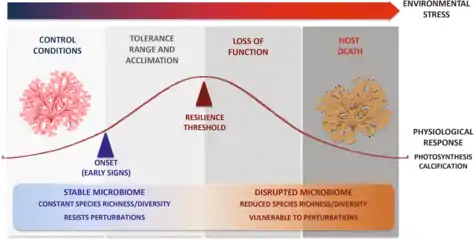
Effects of stressors
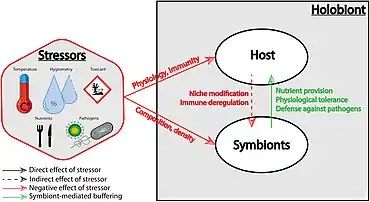
(1) the host physiology and immunity
(2) the symbiotic community composition and density. Stressors may also indirectly affect the symbiotic community by altering the host physiology (which represents the symbiotic niche), and the immune state of the host. Conversely, symbionts can buffer stressors via nutrient provision, physiological tolerance, and defense against host natural enemies.[29]
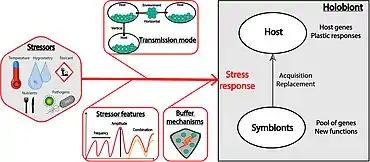
(1) the features of the stressor, such as its frequency or amplitude, but also its combination with another stressor that can lead to an additive, synergistic or antagonist interaction
(2) the transmission mode of the symbionts
(3) the specificity and the efficiency of the given buffering mechanism, and the net balance between its cost and its benefit.[29]
Controversy
Recent years have seen the development of powerful but relatively inexpensive tools for characterising microbial communities, including high throughput sequencing technologies such as whole genome shotgun sequencing. These technological advances have resulted in an explosion of interest in microbial ecology and in the evolution of microbe-host relationships. Some researchers question whether the holobiont concept is needed, and whether it does justice to the intricacies of host-symbiont relationships.[30] In 2016 Douglas and Werren took issue with the concept that "the holobiont (host plus its microbiome) and its constituent hologenome (the totality of genomes in the holobiont) are a unit of selection, and therefore this unit has properties similar to an individual organism".[31] They argue that "the hologenome concept is unhelpful to the study of host interactions with resident microorganisms because it focuses on one level of selection (the holobiont), and as a result it is concerned with cooperative and integrative features of host-microbe systems to the exclusion of other kinds of interactions, including antagonism among microorganisms and conflicts between host and microbial partners."[31]
The holobiont and by extension the hologenome concept remain controversial, particularly in regard to the host and its microbiome as a single evolutionary unit.[32] In order to validate the holobiont concept from an evolutionary perspective, new theoretical approaches are needed that acknowledge the different levels at which natural selection can operate in the context of microbiome-host interactions. For example, selection could occur at the level of the holobiont when a transgenerational association among specific host and symbiont genotypes can be maintained.[32]
Nevertheless, the holobiont concept has resulted in a shift from the focus on symbioses involving one microbial partner and a single host (squids and luminescent Aliivibrio, legumes and Rhizobium, aphids and Buchnera) toward a greater interest in symbioses in complex multi-partner consortia (animal gut systems, marine invertebrates, plant and seaweed epiphytes, microbe-microbe interactions in soil, aquatic biomes).[32] Moreover, there is a realization that even the relatively well understood binary symbioses such as aphids and Buchnera are more complex with a number of diverse facultative symbionts contributing to resistance to parasites,[33] expanding host plant usage[34] and temperature adaptation.[35][32]
References
- Margulis L, Fester R (1991). Symbiosis as a Source of Evolutionary Innovation. MIT Press. ISBN 9780262132695.
- {{cite journal | vauthors = Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TC, Cryan JF, Gilbert SF, Goodnight CJ, Lloyd EA, Sapp J, Vandenkoornhuyse P, Zilber-Rosenberg I, Rosenberg E, Bordenstein SR | title = Getting the Hologenome Concept Right: an Eco-Evolutionary Framework for Hosts and Their Microbiomes | journal = mSystems | volume = 1 | issue = 2 | pages = | date = 2016 | pmid = 27822520 | doi = 10.1128/mSystems.00028-16
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - De Weger LA, van der Vlugt CI, Wijfjes AH, Bakker PA, Schippers B, Lugtenberg B (June 1987). "Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots". Journal of Bacteriology. 169 (6): 2769–73. doi:10.1128/jb.169.6.2769-2773.1987. PMID 3294806.
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A (June 2015). "The importance of the microbiome of the plant holobiont". The New Phytologist. 206 (4): 1196–206. doi:10.1111/nph.13312. PMID 25655016.
- Knowlton N, Rohwer F (October 2003). "Multispecies microbial mutualisms on coral reefs: the host as a habitat". The American Naturalist. 162 (4 Suppl): S51–62. doi:10.1086/378684. PMID 14583857.
- Kramer P, Bressan P (July 2015). "Humans as Superorganisms: How Microbes, Viruses, Imprinted Genes, and Other Selfish Entities Shape Our Behavior". Perspectives on Psychological Science. 10 (4): 464–81. doi:10.1177/1745691615583131. PMID 26177948. S2CID 12954636.
- Sánchez-Cañizares C, Jorrín B, Poole PS, Tkacz A (August 2017). "Understanding the holobiont: the interdependence of plants and their microbiome". Current Opinion in Microbiology. 38: 188–196. doi:10.1016/j.mib.2017.07.001. PMID 28732267.
- Bordenstein SR, Theis KR (August 2015). "Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes". PLoS Biology. 13 (8): e1002226. doi:10.1371/journal.pbio.1002226. PMC 4540581. PMID 26284777.
- Gordon J, Knowlton N, Relman DA, Rohwer F, Youle M (2013). "Superorganisms and holobionts". Microbe. 8 (4): 152–153. doi:10.1128/microbe.8.152.1.
- Wheeler WM (1928). The Social Insects, Their Origin and Evolution. Harcourt Brace.
- Rosenberg E, Zilber-Rosenberg I (March 2016). "Microbes Drive Evolution of Animals and Plants: the Hologenome Concept". mBio. 7 (2): e01395. doi:10.1128/mBio.01395-15. PMID 27034283.
- Ugarelli K, Chakrabarti S, Laas P, Stingl U (December 2017). "The Seagrass Holobiont and Its Microbiome". Microorganisms. 5 (4). doi:10.3390/microorganisms5040081. PMID 29244764.
 Material was copied from this source, which is available under a [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License
Material was copied from this source, which is available under a [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License - van de Guchte M, Blottière HM, Doré J (May 2018). "Humans as holobionts: implications for prevention and therapy". Microbiome. 6 (1): 81. doi:10.1186/s40168-018-0466-8. PMID 29716650.
- Hacquard S, Schadt CW (March 2015). "Towards a holistic understanding of the beneficial interactions across the Populus microbiome". The New Phytologist. 205 (4): 1424–30. doi:10.1111/nph.13133. PMID 25422041.
- Hassani MA, Durán P, Hacquard S (March 2018). "Microbial interactions within the plant holobiont". Microbiome. 6 (1): 58. doi:10.1186/s40168-018-0445-0. PMID 29587885.
- Grasis JA (2017). "The Intra-Dependence of Viruses and the Holobiont". Frontiers in Immunology. 8: 1501. doi:10.3389/fimmu.2017.01501. PMID 29170664.
- Postler TS, Ghosh S (July 2017). "Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System". Cell Metabolism. 26 (1): 110–130. doi:10.1016/j.cmet.2017.05.008. PMID 28625867.
- Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR (November 2016). "Phylosymbiosis: Relationships and Functional Effects of Microbial Communities across Host Evolutionary History". Plos Biology. 14 (11): e2000225. doi:10.1371/journal.pbio.2000225. PMID 27861590.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Cúcio C, Engelen AH, Costa R, Muyzer G (2016). "Rhizosphere Microbiomes of European + Seagrasses Are Selected by the Plant, But Are Not Species Specific". Frontiers in Microbiology. 7: 440. doi:10.3389/fmicb.2016.00440. PMID 27065991.
- Rosenberg E, Zilber-Rosenberg I (April 2018). "The hologenome concept of evolution after 10 years". Microbiome. 6 (1): 78. doi:10.1186/s40168-018-0457-9. PMC 5922317. PMID 29695294.
- Abdelfattah A, Wisniewski M, Schena L, Tack AJ (January 2021). "Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root". Environmental Microbiology. n/a (n/a). doi:10.1111/1462-2920.15392. PMID 33427409.
- Begum N, Qin C, Ahanger MA, Raza S, Khan MI, Ashraf M, Ahmed N, Zhang L (2019). "Role of Arbuscular Mycorrhizal Fungi in Plant Growth Regulation: Implications in Abiotic Stress Tolerance". Frontiers in Plant Science. 10: 1068. doi:10.3389/fpls.2019.01068. PMID 31608075.
- Guerre P (March 2015). "Ergot alkaloids produced by endophytic fungi of the genus Epichloë". Toxins. 7 (3): 773–90. doi:10.3390/toxins7030773. PMID 25756954.
- Schwelm A, Badstöber J, Bulman S, Desoignies N, Etemadi M, Falloon RE, et al. (April 2018). "Not in your usual Top 10: protists that infect plants and algae". Molecular Plant Pathology. 19 (4): 1029–1044. doi:10.1111/mpp.12580. PMID 29024322.
- Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, et al. (November 2018). "Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny". Nature Communications. 9 (1): 4921. doi:10.1038/s41467-018-07275-x. PMC 6250698. PMID 30467310.
- Thompson JR, Rivera HE, Closek CJ, Medina M (2014). "Microbes in the coral holobiont: partners through evolution, development, and ecological interactions". Frontiers in Cellular and Infection Microbiology. 4: 176. doi:10.3389/fcimb.2014.00176. PMID 25621279.
- Pita L, Rix L, Slaby BM, Franke A, Hentschel U (March 2018). "The sponge holobiont in a changing ocean: from microbes to ecosystems". Microbiome. 6 (1): 46. doi:10.1186/s40168-018-0428-1. PMID 29523192.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Cavalcanti GS, Shukla P, Morris M, Ribeiro B, Foley M, Doane MP, Thompson CC, Edwards MS, Dinsdale EA, Thompson FL (September 2018). "Rhodoliths holobionts in a changing ocean: host-microbes interactions mediate coralline algae resilience under ocean acidification". BMC Genomics. 19 (1): 701. doi:10.1186/s12864-018-5064-4. PMID 30249182.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Bénard A, Vavre F, Kremer N (2020). "Stress & Symbiosis: Heads or Tails?". Frontiers in Ecology and Evolution. 8: 167. doi:10.3389/fevo.2020.00167..
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Moran NA, Sloan DB (December 2015). "The Hologenome Concept: Helpful or Hollow?". PLoS Biology. 13 (12): e1002311. doi:10.1371/journal.pbio.1002311. PMC 4670207. PMID 26636661.
- Douglas AE, Werren JH (March 2016). "Holes in the Hologenome: Why Host-Microbe Symbioses Are Not Holobionts". mBio. 7 (2): e02099. doi:10.1128/mBio.02099-15. PMID 27034285.
- Egan S, Fukatsu T, Francino MP (2020). "Opportunities and Challenges to Microbial Symbiosis Research in the Microbiome Era". Frontiers in Microbiology. 11: 1150. doi:10.3389/fmicb.2020.01150. PMID 32612581.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Oliver KM, Russell JA, Moran NA, Hunter MS (February 2003). "Facultative bacterial symbionts in aphids confer resistance to parasitic wasps". Proceedings of the National Academy of Sciences of the United States of America. 100 (4): 1803–7. doi:10.1073/pnas.0335320100. PMID 12563031.
- Tsuchida T, Koga R, Fukatsu T (March 2004). "Host plant specialization governed by facultative symbiont". Science. New York, N.Y. 303 (5666): 1989. doi:10.1126/science.1094611. PMID 15044797.
- Montllor CB, Maxmen A, Purcell AH (2002). "Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress"". Ecological Entomology. 27: 189–195. doi:10.1046/j.1365-2311.2002.00393.x.