LINE1
LINE1 (also L1 and LINE-1) are class I transposable elements in the DNA of some organisms and belong to the group of long interspersed nuclear elements (LINEs). L1 comprise approximately 17% of the human genome.[1] The majority of L1 in the human genome are inactive; however, about 80-100 have retained the ability to retrotranspose, with considerable variation between individuals.[2][3][4] These active L1s can interrupt the genome through insertions, deletions, rearrangements, and copy number variations (CNV).[5] L1 activity has contributed to the instability and evolution of genomes, and is tightly regulated in the germline by DNA methylation, histone modifications, and piRNA.[6] L1s can further impact genome variation by mispairing and unequal crossing-over during meiosis due to its repetitive DNA sequences.[5]
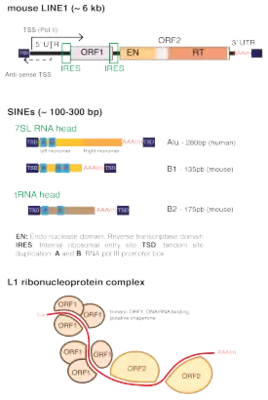
L1 gene products are also required by many nonautonomous Alu and SVA SINE retrotransposons. Mutations induced by L1 and its nonautonomous counterparts have been found to cause a variety of heritable and somatic diseases.[7][8]
Human L1 has been reported to have transferred to the genome of the gonorrhea bacteria.[9][10]
Structure
A typical L1 element is approximately 6,000 base pairs (bp) long and consists of two non-overlapping open reading frames (ORF) which are flanked by untranslated regions (UTR) and target site duplications. In humans, ORF2 is thought to be translated by an unconventional termination/reinitiation mechanism,[11] while mouse L1s contain an internal ribosome entry site (IRES) upstream of each ORF.[12]
5' UTR
The 5' UTR of the L1 element contains a strong, internal RNA Polymerase II transcription promoter in sense[13]
The 5' UTR of mouse L1s contain a variable number of GC-rich tandemly repeated monomers of around 200bp, followed by a short non-monomeric region.
Human 5’ UTRs are ~900bp in length and do not contain repeated motifs. All families of human L1s harbor in their most 5’ extremity a binding motif for the transcription factor YY1.[14] Younger families have also two binding sites for SOX-family transcription factors, and both YY1 and SOX sites were shown to be required for human L1 transcription initiation and activation.[15][16]
Both mouse and human 5’ UTRs contain as well a weak antisense promoter of unknown function.[17][18]
ORF1
| LINE-1 (L1.2) retrotransposable element ORF1 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | L1RE1 | ||||||
| Alt. symbols | L1ORF1p | ||||||
| NCBI gene | 4029 | ||||||
| HGNC | 6686 | ||||||
| OMIM | 151626 | ||||||
| PDB | 2LDY | ||||||
| UniProt | Q9UN81 | ||||||
| Other data | |||||||
| Locus | Chr. 22 q12.1 | ||||||
| Wikidata | Q18028646 | ||||||
| |||||||
The first ORF encode a 500 amino acid - 40 kDa protein that lacks homology with any protein of known function. In vertebrates, it contains a conserved C-terminus domain and a highly variable coiled-coil N-terminus that mediates the formation of ORF1 trimeric complexes. ORF1 trimers have RNA-binding and nucleic acid chaperone activity that are necessary for retrotransposition.[19]
ORF2
| LINE-1 retrotransposable element ORF2 | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | L1RE2 | ||||||
| Alt. symbols | L1ORF2p | ||||||
| NCBI gene | 4030 | ||||||
| HGNC | 6687 | ||||||
| PDB | 1VYB | ||||||
| UniProt | O00370 | ||||||
| Other data | |||||||
| Locus | Chr. 1 q | ||||||
| Wikidata | Q18028649 | ||||||
| |||||||
The second ORF of L1 encodes a protein that has endonuclease and reverse transcriptase activity. The encoded protein has a molecular weight of 150 kDA. The massive antisense expression of RT domain of ORF2 has been reported in primitive eukaryote Entamoeba histolytica which gives the plausible reason of not detecting the ORF2p in this organism [20]
Roles in disease
Cancer
L1 activity has been observed in numerous types of cancers, with particularly extensive insertions found in colorectal and lung cancers.[21] It is currently unclear if these insertions are causal or secondary effects of cancer progression. However, at least two cases have found somatic L1 insertions causative of cancer by disrupting the coding sequences of genes APC and PTEN in colon and endometrial cancer, respectively.[5]
Quantification of L1 copy number by qPCR or L1 methylation levels with bisulfite sequencing are used as diagnostic biomarkers in some types of cancers. L1 hypomethylation of colon tumor samples is correlated with cancer stage progression.[22][23] Furthermore, less invasive blood assays for L1 copy number or methylation levels are indicative of breast or bladder cancer progression and may serve as methods for early detection.[24][25]
Neuropsychiatric disorders
Higher L1 copy numbers have been observed in the human brain compared to other organs.[26][27] Studies of animal models and human cell lines have shown that L1s become active in neural progenitor cells (NPCs), and that experimental deregulation of or overexpression of L1 increases somatic mosaicism. This phenomenon is negatively regulated by Sox2, which is downregulated in NPCs, and by MeCP2 and methylation of the L1 5' UTR.[3] Human cell lines modeling the neurological disorder Rett syndrome, which carry MeCP2 mutations, exhibit increased L1 transposition, suggesting a link between L1 activity and neurological disorders.[28][3] Current studies are aimed at investigating the potential roles of L1 activity in various neuropsychiatric disorders including schizophrenia, autism spectrum disorders, epilepsy, bipolar disorder, Tourette syndrome, and drug addiction.
Retinal disease
Increased RNA levels of Alu, which requires L1 proteins, are associated with a form of age-related macular degeneration, a neurological disorder of the eyes.[29]
The naturally occurring mouse retinal degeneration model rd7 is caused by an L1 insertion in the Nr2e3 gene.[30]
See also
- L1Base, a database of functional annotations and predictions of active LINE1 elements
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (International Human Genome Sequencing Consortium) (February 2001). "Initial sequencing and analysis of the human genome". Nature. 409 (6822): 860–921. Bibcode:2001Natur.409..860L. doi:10.1038/35057062. PMID 11237011.
- Ostertag EM, Kazazian HH (December 2001). "Biology of mammalian L1 retrotransposons". Annual Review of Genetics. 35 (1): 501–38. doi:10.1146/annurev.genet.35.102401.091032. PMID 11700292.
- Erwin JA, Marchetto MC, Gage FH (August 2014). "Mobile DNA elements in the generation of diversity and complexity in the brain". Nature Reviews. Neuroscience. 15 (8): 497–506. doi:10.1038/nrn3730. PMC 4443810. PMID 25005482.
- Hancks DC, Kazazian HH (2016-05-06). "Roles for retrotransposon insertions in human disease". Mobile DNA. 7: 9. doi:10.1186/s13100-016-0065-9. PMC 4859970. PMID 27158268.
- Kazazian HH, Moran JV (July 2017). "Mobile DNA in Health and Disease". The New England Journal of Medicine. 377 (4): 361–370. doi:10.1056/NEJMra1510092. PMC 5980640. PMID 28745987.
- Wang PJ (July 2017). "Tracking LINE1 retrotransposition in the germline". Proceedings of the National Academy of Sciences of the United States of America. 114 (28): 7194–7196. doi:10.1073/pnas.1709067114. PMC 5514774. PMID 28663337.
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV (2011). "LINE-1 elements in structural variation and disease". Annual Review of Genomics and Human Genetics. 12 (1): 187–215. doi:10.1146/annurev-genom-082509-141802. PMC 4124830. PMID 21801021.
- Wimmer K, Callens T, Wernstedt A, Messiaen L (November 2011). "The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion". PLOS Genetics. 7 (11): e1002371. doi:10.1371/journal.pgen.1002371. PMC 3219598. PMID 22125493.
- Yong E (2011-02-16). "Gonorrhea has picked up human DNA (and that's just the beginning)". National Geographic. Retrieved 2016-07-14.
- Anderson, MT; Seifert, HS (2011). "Opportunity and means: horizontal gene transfer from the human host to a bacterial pathogen". mBio. 2 (1): e00005-11. doi:10.1128/mBio.00005-11. PMC 3042738. PMID 21325040.
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV (January 2006). "Unconventional translation of mammalian LINE-1 retrotransposons". Genes & Development. 20 (2): 210–24. doi:10.1101/gad.1380406. PMC 1356112. PMID 16418485.
- Li PW, Li J, Timmerman SL, Krushel LA, Martin SL (2006-01-01). "The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition". Nucleic Acids Research. 34 (3): 853–64. doi:10.1093/nar/gkj490. PMC 1361618. PMID 16464823.
- Swergold GD (December 1990). "Identification, characterization, and cell specificity of a human LINE-1 promoter". Molecular and Cellular Biology. 10 (12): 6718–29. doi:10.1128/MCB.10.12.6718. PMC 362950. PMID 1701022.
- Becker KG, Swergold GD, Ozato K, Thayer RE (October 1993). "Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element". Human Molecular Genetics. 2 (10): 1697–702. doi:10.1093/hmg/2.10.1697. PMID 8268924.
- Tchénio T, Casella JF, Heidmann T (January 2000). "Members of the SRY family regulate the human LINE retrotransposons". Nucleic Acids Research. 28 (2): 411–5. doi:10.1093/nar/28.2.411. PMC 102531. PMID 10606637.
- Athanikar JN, Badge RM, Moran JV (2004-01-01). "A YY1-binding site is required for accurate human LINE-1 transcription initiation". Nucleic Acids Research. 32 (13): 3846–55. doi:10.1093/nar/gkh698. PMC 506791. PMID 15272086.
- Li J, Kannan M, Trivett AL, Liao H, Wu X, Akagi K, Symer DE (April 2014). "An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition". Nucleic Acids Research. 42 (7): 4546–62. doi:10.1093/nar/gku091. PMC 3985663. PMID 24493738.
- Mätlik K, Redik K, Speek M (2006). "L1 antisense promoter drives tissue-specific transcription of human genes". Journal of Biomedicine & Biotechnology. 2006 (1): 71753. doi:10.1155/JBB/2006/71753. PMC 1559930. PMID 16877819.
- Martin SL (2006). "The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition". Journal of Biomedicine & Biotechnology. 2006 (1): 45621. doi:10.1155/jbb/2006/45621. PMC 1510943. PMID 16877816.
- Kaur D, Agrahari M, Singh SS, Mandal PK, Bhattacharya A. "Transcriptomic analysis of Entamoeba histolytica reveals domain-specific sense strand expression of LINE-encoded ORFs with massive antisense expression of RT domain." Plasmid. 2021 Jan 19:102560. doi: 10.1016/j.plasmid.2021.102560. Epub ahead of print. PMID: 33482228.
- Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, et al. (ICGC Breast Cancer Group; ICGC Bone Cancer Group; ICGC Prostate Cancer Group) (August 2014). "Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes". Science. 345 (6196): 1251343. doi:10.1126/science.1251343. PMC 4380235. PMID 25082706.
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, et al. (December 2008). "A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer". Journal of the National Cancer Institute. 100 (23): 1734–8. doi:10.1093/jnci/djn359. PMC 2639290. PMID 19033568.
- Sunami E, de Maat M, Vu A, Turner RR, Hoon DS (April 2011). "LINE-1 hypomethylation during primary colon cancer progression". PLOS ONE. 6 (4): e18884. Bibcode:2011PLoSO...618884S. doi:10.1371/journal.pone.0018884. PMC 3077413. PMID 21533144.
- Sunami E, Vu AT, Nguyen SL, Giuliano AE, Hoon DS (August 2008). "Quantification of LINE1 in circulating DNA as a molecular biomarker of breast cancer". Annals of the New York Academy of Sciences. 1137 (1): 171–4. Bibcode:2008NYASA1137..171S. doi:10.1196/annals.1448.011. PMID 18837943.
- Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, et al. (March 2010). "Implications of LINE1 methylation for bladder cancer risk in women". Clinical Cancer Research. 16 (5): 1682–9. doi:10.1158/1078-0432.CCR-09-2983. PMC 2831156. PMID 20179218.
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. (August 2009). "L1 retrotransposition in human neural progenitor cells". Nature. 460 (7259): 1127–31. Bibcode:2009Natur.460.1127C. doi:10.1038/nature08248. PMC 2909034. PMID 19657334.
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, et al. (November 2013). "Mosaic copy number variation in human neurons". Science. 342 (6158): 632–7. Bibcode:2013Sci...342..632M. doi:10.1126/science.1243472. PMC 3975283. PMID 24179226.
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH (November 2010). "L1 retrotransposition in neurons is modulated by MeCP2". Nature. 468 (7322): 443–6. Bibcode:2010Natur.468..443M. doi:10.1038/nature09544. PMC 3059197. PMID 21085180.
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, et al. (March 2011). "DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration". Nature. 471 (7338): 325–30. Bibcode:2011Natur.471..325K. doi:10.1038/nature09830. PMC 3077055. PMID 21297615.
- Chen J, Rattner A, Nathans J (July 2006). "Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements". Human Molecular Genetics. 15 (13): 2146–56. doi:10.1093/hmg/ddl138. PMID 16723373.