Symbiosome
A symbiosome is a specialised compartment in a host cell that houses an endosymbiont in a symbiotic relationship.[1]
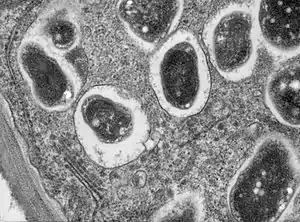
The term was first used in 1983 to describe the vacuole structure in the symbiosis between the animal host the Hydra, and the endosymbiont Chlorella. Symbiosomes are also seen in other cnidaria-dinoflagellate symbioses, including those found in coral-algal symbioses. In 1989 the concept was applied to the similar structure found in the nitrogen-fixing root nodules of certain plants.[1]
The symbiosome in the root nodules has been much more successfully researched due in part to the complexity of isolating the symbiosome membrane in animal hosts.[1] The symbiosome in a root nodule cell in a plant is an organelle-like structure that has formed in a symbiotic relationship with nitrogen-fixing bacteria. The plant symbiosome is unique to those plants that produce root nodules.[2] The majority of such symbioses are made between legumes and diazotrophic Rhizobia bacteria. The rhizobia-legume symbioses are the most studied due to the importance in agriculture.[3][4]
Each symbiosome in a root nodule cell encloses a single rhizobium that differentiates into a bacteroid. However, in some cases a symbiosome may house several bacteroids.[5] The symbiosome membrane, or peribacteroid membrane, surrounds the bacteroid membrane, separated by a symbiosome space. This unit provides an inter-kingdom, micro-environment for the production of nitrogen for the plant,[3][6] and the receipt of malate for energy for the bacteroid.[7]
History
The concept of the symbiosome was first described in 1983, by Neckelmann and Muscatine, as seen in the symbiotic relationship between Chlorella ( a class of green algae, and Hydra a cnidarian animal host.[1] Until then it had been described as a vacuole. A few years later in 1989, Lauren Roth with Gary Stacey [8] as well as Robert B Mellor [9] applied this concept to the nitrogen-fixing unit seen in the plant root nodule,[1] previously called an infection vacuole.[10]
This has since engendered a great deal of research, one result of this has been the provision of a more detailed description of the symbiosome (peribacteroid) membrane, as well as comparisons with similar structures in Vesicular Arbuscular Mycorrhizal symbioses in plants.[11] In the animal models, the symbiosome has a more complex arrangement of membranes, such that it has proved difficult to isolate, purify and study.[1]
Structure and formation
A symbiosome is formed as a result of a complex and coordinated interaction between the symbiont host and the endosymbiont.[5] At the point of entry into a symbiont host cell, part of the cell's membrane envelops the endosymbiont and breaks off into the cytoplasm as a discrete unit, an organelle-like vacuole called the symbiosome.[5][12] This is an endocytosis-like process that forms a symbiosome rather than an endosome. In plants this process is unique.[13]
The symbiosome membrane is separated from the endosymbiont membrane by a space known as the symbiosome space, which allows for the exchange of solutes between the symbionts.[14][12] In the plant root nodule the symbiosome membrane is also called the peribacteroid membrane.[13]
In the plant
In the legume-rhizobia symbioses the symbiosome is the nitrogen-fixing unit in the plant, formed by an interaction of plant and bacterial signals, and their cooperation. The legumes are protein-rich, and have a high demand for nitrogen that is usually available from nitrates in the soil. When these are scarce the plant secretes flavonoids that attract free-living diazotrophic (nitrogen-fixing) rhizobia to their root hairs. In turn the bacteria release Nod factors that stimulate the infection process in the plant.[1][13]
To enable infection the tip of the root hair curls over the rhizobia and by an inward growth produces an infection thread to carry the endosymbionts into the cortical cells. At the same time the cortical cells divide to produce the tough root nodules that will house and protect the bacteria.[15][13] The bacterial production of extracellular polymeric substance (EPS) is seen to be necessary for enabling infection.[13] The rhizobia infect the plant in large numbers, only seen to be actively dividing at the tip of the injection thread, where they are released into the cells inside symbiosomes.[15][1] The symbiosome is formed as a result of an endocytosis-like process that produces an endosome. Typically endosomes target to lysosomes, but the symbiosome re-targets the host-cell proteins.
The changes in the plant needed to form the infection thread, the increased division of the cortical cells, the formation of the root nodule, and symbiosome, are brought about by dynamic changes in the actin cytoskeleton.[16][13] Filamentous actin (F-actin) channels the elongation of the injection threads and short F-actin fragments are dotted around the symbiosome membrane.[16] The bacteria are released as injection drops into the host root nodule cells where the plasma membrane encloses them in the organelle-like structure of the symbiosome. In most plants a symbiosome encloses a single endosymbiont bacterium but some types may contain more than one. A negative feedback loop called the autoregulation of nodulation works to balance the need for nitrogen and thus the formation of nodules.[17][18]
Differentiation
The outer host-cell derived symbiosome membrane encloses a space called the symbisome space or the peribacteroid space that surrounds the endosymbiont. In order for the symbiosome to be established as a nitrogen-fixing unit the enclosed bacterium has to be terminally differentiated into a morphologically changed bacteroid. The bacterium in the soil is free-living and motile. In the symbiosome it has to change its gene expression to adapt to a non-motile, non-reproductive form as the bacteroid. This change is noted by an increase in the size of the bacterium and its elongation. The bacterial membrane is also made permeable.[19][1][13] The process of differentiation is plant-driven using peptides known as nodule specific cysteine-rich peptides (NCR peptides).
NCRs are antimicrobial peptides that are similar to the defensin peptides used in mammals in response to invading pathogens. The NCRs are targeted to the symbiosome where they induce differentiation of the bacterium to the bacteroid. A major effect of NCR targeting is to limit the reproductive ability of the endosymbiont. These changes are controlled, since the bacterium is not killed as a result of exposure to the NCRs. Some of that control comes from the bacterium itself.[20][21][5] In order to survive the NCR activities, the bacteria need to produce a protein called BacA. In addition the lipopolysaccharide produced by the bacteria is modified by an unusual fatty acid that also gives protection against environmental stresses. These defensive measures help the differentiation process and ensures their survival as bacteroids. Some strains of rhizobia produce a peptidase that degrades the NCRs.[20][21]
Nitrogen-fixing unit
The established bacteroid is able to fix nitrogen into a chemically usable form of ammonium for the plant. This is an energy-demanding process fuelled by the plant's carbohydrates.[13] Transport vesicles form in the symbiosome membrane allowing the passage of ammonium into the symbiosome space from the bacteroid, and the passage of plant nutrients to the bacteroid.[13] The rhizobia infect the plant in large numbers where they are released into the cells inside symbiosomes. They are protected by the tough structure of the root nodule.[15]
In the animal
The most well studied symbiosis involving an animal host is that between the cnidaria and the dinoflagellates, most commonly the single-celled zooxanthellae. The symbiosis of the Chlorella–Hydra first described the symbiosome. The coral Zoanthus robustus has been used as a model organism to study the symbiosis with its microsymbiont algal species of Symbiodinium, with a focus on the symbiosome and its membranes. Methods for isolating the symbiosome membranes have been looked for – the symbiont in the animal host has a multilayered membrane complex which has proved resistant to disruption making their isolation difficult.[1][22]
The endosymbiont dinoflagellates are used for their ability to photosynthesise and provide energy, giving the host cnidarians such as corals, and anemones, plant properties.[23] Free-living dinoflagellates are ingested into the gastrodermal cells of the host, and their symbiosome membrane is derived from the host cell.[24] The process of symbiosome formation is often seen in the animal host to be that of phagocytosis,[24] and it is hypothesised that the symbiosome is a phagosome that has been subject to early arrest.[25]
Similar structures
A similar structure to the symbiosome is the parasitophorous vacuole formed within host cells infected by apicomplexan parasites. The vacuole is derived from the host cell plasma membrane. It is made safe from the host's endolysomal system by modifying-proteins released by the parasite.[26][27] The parasitophorous vacuole membrane is greatly remodelled by the parasite.[28]
See also
References
- "(PDF) Isolation of Symbiosomes and The Symbiosome Membrane Complex from The Zoanthid Zoanthus Robustus". ResearchGate.
- Emerich, DW; Krishnan, HB (15 May 2014). "Symbiosomes: temporary moonlighting organelles". The Biochemical Journal. 460 (1): 1–11. doi:10.1042/BJ20130271. PMID 24762136.
- Coba de la Peña, T; Fedorova, E; Pueyo, JJ; Lucas, MM (2017). "The Symbiosome: Legume and Rhizobia Co-evolution toward a Nitrogen-Fixing Organelle?". Frontiers in Plant Science. 8: 2229. doi:10.3389/fpls.2017.02229. PMC 5786577. PMID 29403508.
- Zahran, HH (December 1999). "Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate". Microbiology and Molecular Biology Reviews. 63 (4): 968–89, table of contents. doi:10.1128/mmbr.63.4.968-989.1999. PMC 98982. PMID 10585971.
- Haag, AF; et al. (May 2013). "Molecular insights into bacteroid development during Rhizobium-legume symbiosis". FEMS Microbiology Reviews. 37 (3): 364–83. doi:10.1111/1574-6976.12003. PMID 22998605.
- Andrews, M; Andrews, ME (26 March 2017). "Specificity in Legume-Rhizobia Symbioses". International Journal of Molecular Sciences. 18 (4): 705. doi:10.3390/ijms18040705. PMC 5412291. PMID 28346361.
- Schulze, J.; et al. (1 November 2002). "Malate plays a central role in plant nutrition". Plant and Soil. 247: 133–139. doi:10.1023/A:1021171417525.
- Roth, LE; Stacey, G (June 1989). "Bacterium release into host cells of nitrogen-fixing soybean nodules: the symbiosome membrane comes from three sources". European Journal of Cell Biology. 49 (1): 13–23. PMID 2759097.
- Mellor, RB (June 1989). "Bacteroids in the Rhizobium-legume symbiosis inhabit a plant internal lytic compartment: implications for other microbial endosymbioses". Journal of Experimental Botany. 40 (3): 831–839.
- Goodchild, DJ; Bergersen, FJ (July 1966). "Electron microscopy of the infection and subsequent development of soybean nodule cells". Journal of Bacteriology. 92 (1): 204–13. doi:10.1128/jb.92.1.204-213.1966. PMC 276217. PMID 5949564.
- Mellor, RB; et, al (May 1990). "Vesicular-arbuscular mycorrhizas of wild-type soybean and non-nodulating mutants with Glomus mosseae contain symbiosis-specific polypeptides (mycorrhizins), immunologically cross-reactive with nodulins". Planta. 182 (1): 22–26.
- Kereszt, A; Mergaert, P; Kondorosi, E (November 2011). "Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims?". Molecular Plant-Microbe Interactions. 24 (11): 1300–9. doi:10.1094/MPMI-06-11-0152. PMID 21995798.
- Long, SR (6 October 2016). "SnapShot: Signaling in Symbiosis". Cell. 167 (2): 582–582.e1. doi:10.1016/j.cell.2016.09.046. PMID 27716511.
- Mouritzen, P; Rosendahl, L (October 1997). "Identification of a Transport Mechanism for NH4+ in the Symbiosome Membrane of Pea Root Nodules". Plant Physiology. 115 (2): 519–526. doi:10.1104/pp.115.2.519. PMC 158510. PMID 12223820.
- Buhian, WP; Bensmihen, S (2018). "Mini-Review: Nod Factor Regulation of Phytohormone Signaling and Homeostasis During Rhizobia-Legume Symbiosis". Frontiers in Plant Science. 9: 1247. doi:10.3389/fpls.2018.01247. PMC 6166096. PMID 30319665.
- Zhang, X; Han, L; Wang, Q; Zhang, C; Yu, Y; Tian, J; Kong, Z (January 2019). "The host actin cytoskeleton channels rhizobia release and facilitates symbiosome accommodation during nodulation in Medicago truncatula". The New Phytologist. 221 (2): 1049–1059. doi:10.1111/nph.15423. PMID 30156704.
- Wang, C; Reid, JB; Foo, E (2018). "The Art of Self-Control - Autoregulation of Plant-Microbe Symbioses". Frontiers in Plant Science. 9: 988. doi:10.3389/fpls.2018.00988. PMC 6048281. PMID 30042780.
- Reid, DE; Ferguson, BJ; Hayashi, S; Lin, YH; Gresshoff, PM (October 2011). "Molecular mechanisms controlling legume autoregulation of nodulation". Annals of Botany. 108 (5): 789–95. doi:10.1093/aob/mcr205. PMC 3177682. PMID 21856632.
- Alunni, B; Gourion, B (July 2016). "Terminal bacteroid differentiation in the legume-rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond". The New Phytologist. 211 (2): 411–7. doi:10.1111/nph.14025. PMID 27241115.
- Maróti, G; Downie, JA; Kondorosi, É (August 2015). "Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules". Current Opinion in Plant Biology. 26: 57–63. doi:10.1016/j.pbi.2015.05.031. PMID 26116977.
- Pan, H; Wang, D (4 May 2017). "Nodule cysteine-rich peptides maintain a working balance during nitrogen-fixing symbiosis". Nature Plants. 3 (5): 17048. doi:10.1038/nplants.2017.48. PMID 28470183.
- Davy, SK; Allemand, D; Weis, VM (June 2012). "Cell biology of cnidarian-dinoflagellate symbiosis". Microbiology and Molecular Biology Reviews. 76 (2): 229–61. doi:10.1128/MMBR.05014-11. PMC 3372257. PMID 22688813.
- Allemand, D; Furla, P (May 2018). "How does an animal behave like a plant? Physiological and molecular adaptations of zooxanthellae and their hosts to symbiosis". Comptes Rendus Biologies. 341 (5): 276–280. doi:10.1016/j.crvi.2018.03.007. PMID 29650460.
- Peng, SE; et al. (March 2010). "Proteomic analysis of symbiosome membranes in Cnidaria-dinoflagellate endosymbiosis". Proteomics. 10 (5): 1002–16. doi:10.1002/pmic.200900595. PMID 20049864.
- Mohamed, AR; et al. (July 2016). "The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: the symbiosome as an arrested early phagosome". Molecular Ecology. 25 (13): 3127–41. doi:10.1111/mec.13659. PMID 27094992.
- Clough, B; Frickel, EM (June 2017). "The Toxoplasma Parasitophorous Vacuole: An Evolving Host-Parasite Frontier". Trends in Parasitology. 33 (6): 473–488. doi:10.1016/j.pt.2017.02.007. PMID 28330745.
- Lingelbach, K; Joiner, KA (June 1998). "The parasitophorous vacuole membrane surrounding Plasmodium and Toxoplasma: an unusual compartment in infected cells". Journal of Cell Science. 111 ( Pt 11): 1467–75. PMID 9580555.
- Burda, Paul-Christian; Heussler, Volker T.; Brühlmann, Francis; Bausch-Fluck, Damaris; Schnider, Cilly Bernardette (28 February 2018). "BioID Reveals Novel Proteins of the Plasmodium Parasitophorous Vacuole Membrane". mSphere. 3 (1): e00522–17. doi:10.1128/mSphere.00522-17. PMC 5784244. PMID 29404413.