TMEM267
TMEM267 is a protein that in humans is encoded by the TMEM267 gene. It is a possible oncogene which encodes a transmembrane protein. The function of TMEM267 most likely involves transportation of molecules from the cytosol, as the presence of motifs and domains involved in transportation were conserved in orthologs. TMEM267 has orthologs in many species and is expressed at highest levels in the thyroid.
| TMEM267 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | TMEM267, C5orf28, transmembrane protein 267 | ||||||||||||||||||||||||
| External IDs | MGI: 3648543 HomoloGene: 49708 GeneCards: TMEM267 | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) |
| ||||||||||||||||||||||||
| Location (UCSC) | Chr 5: 43.44 – 43.48 Mb | Chr 13: 119.49 – 119.61 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [4] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Gene
Known aliases for TMEM267include C5orf28, B2RDA6, and Q9H6Z2.[5] TMEM267 is found on chromosome 5, cytoband p12 on the reverse strand between base pairs 43,444,252 and 43,485,178, meaning it has length of 40,927 base pairs.[6] TMEM267 produces 13 distinct gt-ag introns and 12 different mRNAs, with 9 alternatively spliced variants and 3 unspliced forms. It has 2 alternative promoters and 7 validated polyadenylation sites.[7] There are 6 predicted promoters of varying lengths.[8]
| Name | Accession Number | Number of Exons | Size (bp) |
|---|---|---|---|
| Transcript Variant 1 | NM_022483 | 3 | 2736 |
| Transcript Variant 2 | NM_001377394.1 | 4 | 2887 |
| Transcript Variant X2 | XM_011514075 | 4 | 3574 |
| Transcript Variant 3 | NM_001377395.1 | 5 | 3210 |
| Transcript Variant 4 | NM_001377396.1 | 3 | 2960 |
| Transcript Variant 5 | NM_001377397.1 | 4 | 3108 |
| Transcript Variant 6 | NM_001377398.1 | 5 | 3283 |
| Transcript Variant 7 | NM_001377399.1 | 3 | 3377 |
| Transcript Variant 8 | NM_001377400.1 | 4 | 3528 |
| Transcript Variant 9 | NM_001377401.1 | 4 | 2834 |
| Transcript Variant 10 | NM_001377402.1 | 3 | 2809 |
| Transcript Variant 11 | NM_001377403.1 | 5 | 2979 |
Protein
General information
The TMEM267 protein in all isoforms is 215 amino acids in length.[9] All of the isoforms have a predicted molecular mass of 24.2 kDa and theoretical isoelectric point of 8.91.[10][11] There was an above average percentage of histidine and tryptophan residues. The percentage of asparagine, glutamic acid and tyrosine were below average. After analysis of Antarctic Yellowbelly Rockcod, Tropical clawed frog, Willow flycatcher, Common wall lizard, and pacific white-sided dolphin orthologs, above average percent composition of tryptophan and below average percent composition of asparagine and glutamic acid residues was conserved across amphibians, fish, mammals, birds, and reptiles.[12]
Isoforms
| Name | Accession Number | Size (aa) |
|---|---|---|
| Transcript Variant 2 | NP_001364323.1 | 215 |
| Transcript Variant X2 | XM_011514075 | 215 |
| Transcript Variant 3 | NP_001364324 | 215 |
| Transcript Variant 4 | NP_001364325 | 215 |
| Transcript Variant 5 | NP_001364326 | 215 |
| Transcript Variant 6 | NP_001364327 | 215 |
| Transcript Variant 7 | NP_001364328 | 215 |
| Transcript Variant 8 | NP_001364329 | 215 |
| Transcript Variant 9 | NP_001364330 | 215 |
| Transcript Variant 10 | NP_001364331 | 215 |
| Transcript Variant 11 | NP_001364332 | 215 |
| Transcript Variant 12 | NP_071928 | 215 |
Transmembrane regions
The predictions surrounding the transmembrane regions of TMEM267 are unclear. Some prediction tools claim there are no transmembrane or hydrophobic regions.[13] Others predict between 2 and 5 transmembrane domains.[14] [15] The consensus is that there is most likely at least two transmembrane regions in the amino acids in the region around 113-135 and 176–198. The helical wheel diagrams of the three transmembrane regions given by NCBI Gene indicate the presence of polar amino acid which are basic and acidic in the transmembrane regions which is unusual.[16]

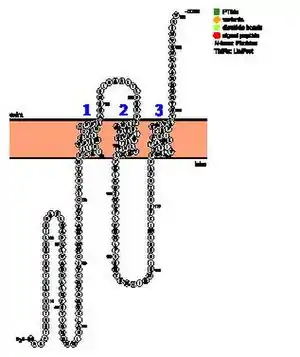
Domains
Two domains were predicted for TMEM267. First is a LaxA-binding, inner membrane-associated mutative hydrolase, which indicates that TMEM267 may be involved as a group of membrane-bound metal-dependent hydrolases that may act as phospholipases. The other is predicted to be a Vacuolar sorting protein 9 (VPS9) domain, which indicates that TMEM267 may be involved in trafficking of molecules to lysosomes and cell signaling.[17][18]
| Domain | Location | Conserved in Isoforms | Conserved in Orthologs |
|---|---|---|---|
| LaxA binding site | 55-161 | yes | yes |
| Vacuolar Sorting Protein 9 (VSP9) binding site | 175-199 | yes | no |
Motifs
Motifs that were predicted are a canonical arginine-containing phosphopeptide motif, which may be involved in the transportation of the 14-3-3 proteins which are involved in many cellular processes.[19] A binding site for Interferon Regulatory Factor 3 (IRF-3) protein may be involved in the signaling pathway which actives IRF-3 in the presence of viral and microbial infections.[20] Tryptophan-based motifs which enable targeting of tethering to a homology domain could mean that TMEM267 may play a role in mediating transportation form the Golgi to the ER. The coatomer subunit delta (delta-COP) is a cytosolic protein complex that binds to motifs and associates with vesicles involved in protein transport from the ER and Golgi.[21] An LC3-Interacting Region (LIR) motif was predicted, which indicates that TMEM267 may be involved in the autophagy pathway, which is involved in transferring of cytoplasmic material in autophagosomes to lysosomes as well as removal of toxic macromolecules and organelles to maintain the health of the cell.[22] The Class 2 PDZ-binding motif predicts that TMEM267 could be involved with PDZ proteins, which are influential in trafficking, recycling, and intracellular sorting.[23] The Wxxx[FY] motif indicates that TMEM267 could be involved in the interaction of Pex14 with Pex5 proteins.[24][25]
| Motif | Location | Conserved in Isoforms | Conserved in Orthologs | Cellular Location |
|---|---|---|---|---|
| Canonical arginine-containing phosphopeptide motif | 49-56 | Yes | Yes | cytosol |
| Interferon Regulatory Factor 3 (IRF-3) binding site | 139-146 | Yes | Yes | cytosol |
| Tryptophan-based motifs that enable tethering to a homology domain | 144-151 | Yes | Yes | cytosol |
| LIR motif | 64-68 | Yes | Yes | cytosol |
| PDZ-binding motif | 210-215 | Yes | Yes | cytosol |
| Wxxx[FY] motif | 166-170 | Yes | Yes | cytosol |
Localization and abundance
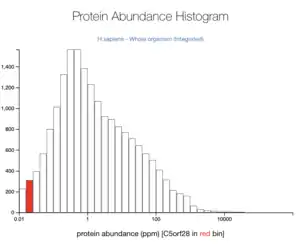
Overall, TMEM267 is most likely found in the cytoplasm. The TMEM267 protein was claimed to be localized in the nucleoplasm of cells.[26] Another tool predicted it to be found in the cytoplasm (69.6%) and mitochondrial (13%) with reliability 94.1 from Reinhardt's method for Cytoplasmic/Nuclear discrimination.[27] TMEM267 is not abundant in the human body at 0.05 ppm.[28]
Secondary structure predictions
Secondary structure predictions were done using the transmembrane regions for TMEM267 given by NCBI Gene. The prediction servers indicate that that amino acids 1-76 are helical in nature. The extracellular and intracellular regions of the TMEM267 protein are predicted to be a combination of alpha helices and beta sheets, but there is not a consensus.[29][30][31]
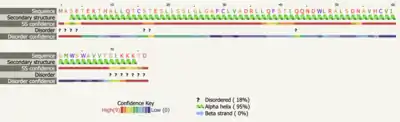
Post-translational modifications
There is no evidence of post-translational modifications of the TMEM267 protein found in tissues.[32] According to protein sequence analysis, there is a prediction of one palmitoylation site, a SUMO Interaction and two sumoylation sites.[33][34] There are many predicted phosphorylation sites in the non-transmembrane regions with various protein kinases including AGC, CKII, and Case kinase II.[35][36] One site is predicted to be acetylated in the N-terminus of TMEM267.[37] TMEM267 has four predicted glycation sites, as well as seven O-beta-GlycNAc sites.[38][39]
Expression
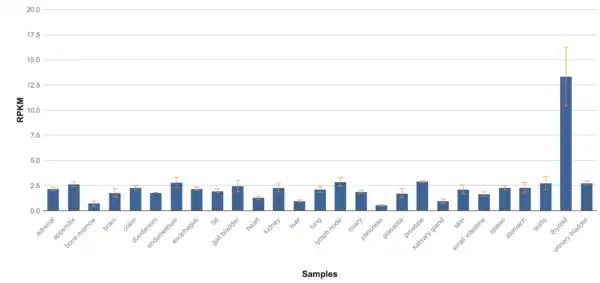
TMEM267 protein is expressed in over 100 tissues in the body, meaning it has low tissue specificity, but is largely present in the thyroid, pituitary gland, and pancreas. Data from NCBI Geo shows that there are higher levels of expression in mainly the thyroid but other tissues have varied expression for each sample. There seems to be, on average, highest expression levels in thyroid, ovaries, testes, pituitary gland, and pancreas. TMEM267 is not expressed at a very high level compared to Beta Actin, which has almost triple the RPKM compared to TMEM267.[40] TMEM267 is expressed at 1.6 times the average gene on chromosome 5.[41]
Interactions
Transcription factors
The table below describes a curated set of transcription factors which are predicted to bind in the Genomatix predicted TMEM267 promoter region.[42]
| Transcription Factor | Detailed information |
|---|---|
| RU49 | Zinc finger proliferation 1-Zipro |
| SORY | SOX-SOY-sex/testis determining and related HMG box factors |
| FKHD | Fork head domain factors |
| BRAC | Brachyury gene, mesoderm developmental factor |
| EVI1 | EVI1-myleoid transformation protein |
| VTBP | Vertebrate TATA binding protein factor |
| RUSH | SWI/SNF related nucelophosphoproteins with RING finger DNA binding motif |
| NKXH | NKX homeodomain factors |
| BRN5 | Brn-5 POU domains |
| PDX1 | Pancreatic and intestinal homeodomain TF |
| CEBPA/B | CCAAT/Enhancer Binding Protein |
| CAAT | CCAAT binding factors |
| SPZ1 | Testis-specific bHLH-Zip TFs |
| NFAT | Nuclear factor of activated T-cells |
| DMRT | DM- domain-containing TFs |
| ZF01 | C2H2 zinc finger TF1 |
| CREB | cAMP-responsive element binding proteins |
| XBBF | X-box binding factors |
| GCNR | Germ cell nuclear receptor |
| PLZF | C2H2 zinc finger protein PLZF |
Interacting proteins
TMEM267 was predicted to interact with the proteins in the table below.
| Protein Name | UniProt Label | Function | Location |
|---|---|---|---|
| TMEM52B | Q4KMG9 | Function has not been studied | Integral component of membrane, extracellular region |
| TMEM14B | Q9NUH8 | Involved in the cortical expansion and folding in the developing neocortex; may drive neural progenitor proliferation through nuclear translocation | Integral component of membrane |
| RTP2 (Receptor-transporting protein 2) | Q5QGT7 | Promotes functional cell surface expression of olfactory receptor | Plasma membrane |
| SAR1A | Q9NR31 | Involved in transport from the ER to the Golgi apparatus; SAR1S-GTP-dependent assembly of SEC16A on the ER membrane form an organized scaffold defining ER exit sites | ER, Golgi apparatus |
| STX7 (Syntaxin-7) | Q15400 | May be involved in protein trafficking from the plasma membrane to early endosome; mediates trafficking from early to late endosomes and lysosomes | Endosome, plasma membrane |
| CPLX4 (Complexin-4) | Q7Z7G2 | Regulates SNARE protein complex-mediated synaptic vesicle fusion | Plasma membrane |
| APP (Amyloid-beta precursor protein P4) | P05067 | Functions as cell surface receptor in neurons; involved in cell mobility and transcription regulation | Extracellular region/secreted, plasma membrane, endosome, nucleus, cytoplasmic vesicle |
| EGFR | P00533 | Ligand binding trigger receptor homo/heterodimerization and autophosphorylation on key cytoplasmic residues; this phosphorylated receptor recruits adaptor proteins which activate downstream signaling cascades | Nuclear membrane, ER membrane |
| LNPEP (Leucyl-cystinyl aminopeptidase) | Q9UIQ6 | Release of an N-terminal amino acid, cleaves before cysteine and leucine; helps maintain homeostasis during pregnancy | Plasma membrane, extracellular region |
| ECM29 | Q5VYK3 | Adaptor/scaffolding protein which binds to specific proteins; may couple the proteasome to ER, endosome, and centrosome | Nucleus, centrosome, ER, endosome, cytoplasmic vesicle |
Homology and evolution
Orthologs and paralogs
TMEM267 has orthologs in Mammalia, Reptilia, Amphibia, Mollusca, Arthropoda, Branchiostoma, Trichoplax, Oomycetes, and Bacteria, among others, but has no paralogs. A table of select orthologs is listed below.[43]
| Genus and Species | Common Name | Taxonomic Group | Estimated Date of Divergence (MYA) | Accession Number | Sequence Length (aa) | Sequence Alignment | Sequence Similarity |
|---|---|---|---|---|---|---|---|
| Nannospalax galili | Northern Israeli Blind Subterranean Mole | Rodentia | 90 | XP_008831143.1 | 215 | 85.19% | 93% |
| Opisthocomus hoazin | Reptile Bird | Opisthocomiformes | 312 | XP_009941974.1 | 215 | 83.26% | 92% |
| Protobothrops mucrosquamatus | Venomous pit viper | Squamata | 312 | XP_015672610.1 | 219 | 82.16% | 89% |
| Xenopus Tropicalis | Tropical clawed frog | Anura | 351.8 | XP_031750178.1 | 215 | 74.88% | 86% |
| Notothenia coriiceps | Antarctic Yellowbelly Rockcod | Perciformes | 435 | XP_010772860.1 | 244 | 67.21% | 82% |
| Branchiostoma belcheri | Branchiostoma | Amphioxiformes | 684 | XP_019622820.1 | 215 | 32.09% | 63% |
| Strongylocentrotus purpuratus | Purple Sea Urchin | Echinodermata | 684 | XP_030828106.1 | 221 | 36.3% | 59% |
| Aethina tumida | Small Hive Beetle | Coleoptera | 797 | XP_019869992.1 | 201 | 35.82% | 49% |
| Crassostrea gigas | Pacific Oyster | Ostreoida | 797 | XP_011423125.1 | 209 | 34.93% | 52% |
Evolution
TMEM267 is predicted to evolve slower than Fibrinogen Alpha Chain but faster than Cytochrome C.[44]
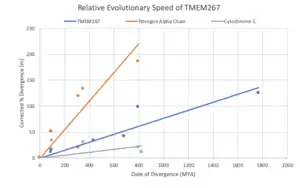
Function
Clinical significance
The protein was identified as a member of a large group of proteins that comprise a filter in mammalian cells which allow selective passage of proteins in and out of the cilium, regulating the contents.[45] TMEM267 was one of ten genes selected using the two-sample t-test and Wilcoxon Mann-Whitney analysis of training data on atopic dermatitis (a skin disease characterized by areas of severe itching, redness, scaling, and loss of the surface of the skin), as a gene that provided the most information about the separation between the control and experimental groups.[46] TMEM267 is mentioned in articles pertaining to the down-regulation of two miRNAs, one of which is involved in regulating a wide variety of cellular functions, such as proliferation, apoptosis, migration, and differentiation, all of which are vital for the normal development of heart cells.[47][48]
Cancer
TMEM267 protein was shown to be mutated in 0.1-0.9% of colorectal, stomach, lung, endometrial, kidney, and breast cancer.[49] TMEM267 was affected by increased levels of the NUDT21 gene, and was identified as a part of a large group of possible oncogenes, that when the 3'-UTR is shortened, can cause uncontrollable cell growth.[50] It is a part of a group of genes which can possibly identify survival rates of patients with PNI+ tongue cancer.[51] TMEM267 was shown to be one of 26 over-expressed genes on chromosome 5p, meaning it belongs to a group of genes which likely provides cancer cells with advantages in growth and invasion of surrounding cells.[52] In addition, researchers from The Johns Hopkins University filed a patent for 63 genes, including TMEM267 which had increased expression in the presence of HMGA1 protein compared to the control group, which they think could be of use in a method of inhibiting cancer stem cells with HMGA1 inhibitors.[53]
References
- GRCh38: Ensembl release 89: ENSG00000151881 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000074634 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- GeneCards.org entry on TMEM267
- GeneCards.org entry on TMEM267
- UniProtKB entry on Q0VDI3 (TM267_HUMAN)
- El Dorado at Genomatix
- NCBI (National Center for Biotechnology Information) AceView entry on C5orf28
- GeneCards.org entry on TMEM267
- ExPASy Compute pI/MW predictor /
- SAPS entry on TMEM267
- SAPS entry on TMEM267
- ELM entry for TMEM267
- Prodom entry on TMEM267
- NetWheels Helical Projection for TMEM267
- MotifFinder entry on TMEM267
- Hislop JN, Marley A, von Zastrow M. Role of Mammalian Vacuolar Protein-sorting Proteins in Endocytic Trafficking of a Non-ubiquitinated G Protein-coupled Receptor to Lysosomes. J Biol Chem 2004;279:22522–22531.
- ELM Detail of Canonical Arginine-containing phosphopeptide motif
- IRF3 information
- delta-COP protein
- Wirth, M., Zhang, W., Razi, M. et al. Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat Commun 10, 2055 (2019).
- Romero, G., von Zastrow, M., & Friedman, P. A. (2011). Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: means, motif, and opportunity. Advances in pharmacology (San Diego, Calif.), 62, 279–314.
- ELM Details for LIG_Pex14
- ELM entry for TMEM267
- The Human Protein Atlas entry on TMEM267
- PSORT prediction for TMEM267
- PaxDb protein abundance
- Chau-Fasman prediction for TMEM267
- GOR4 prediction for TMEM267
- Phyre 2 Prediction for TMEM267
- The Human Protein Atlas entry on TMEM267
- CSS-Palm for TMEM267
- SUMOsp prediction for TMEM267
- GPS for TMEM267
- NetPhos prediction for TMEM267
- NetACET prediction for TMEM267
- NetGlycate prediction for TMEM267
- YinOYang prediction for TMEM267
- NCBI (National Center for Biotechnology Information) AceView entry on C5orf28
- NCBI (National Center for Biotechnology Information) AceView entry on C5orf28
- El Dorado at GenoMatix MatInspector for TMEM267
- NCBI (National Center for Biotechnology Information) AceView entry on C5orf28
- TimeTree Data on Date of Divergence
- Valentine, Megan Smith, "Polycystin-2 (PKD2), Eccentric (XNTA), and Meckelin (MKS3) in the Ciliated Model Organism Paramecium tetraurelia" (2015). Graduate College Dissertations and Theses. 419.
- Tadesse, Dawit. (2018). A Comparison of Selected Parametric and Non-Parametric Statistical Approaches for Candidate Genes Selection in Transcriptome Data.
- Han, S., Wang, W., Duan, L., Hou, Z., Zeng, J., Li, L., … Jiang, L. (2019). MicroRNA profiling of patients with sporadic atrial septal defect. Biotechnology & Biotechnological Equipment, 33(1), 510–519.
- Sun, Xiaoyan & Song, Zhenhua & Si, Yawei & Wang, Jin-Hui. (2018). microRNA and mRNA profiles in ventral tegmental area relevant to stress-induced depression and resilience. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 86. 10.1016
- Yao, Z., Darowski, K., St-Denis, N., Wong, V., Offensperger, F., Villedieu, A., … Stagljar, I. (2017). A Global Analysis of the Receptor Tyrosine Kinase-Protein Phosphatase Interactome. Molecular cell, 65(2), 347–360.
- Xiong, M., Chen, L., Zhou, L., Ding, Y., Kazobinka, G., Chen, Z., & Hou, T. (2019). NUDT21 inhibits bladder cancer progression through ANXA2 and LIMK2 by alternative polyadenylation. Theranostics, 9(24), 7156–7167.
- Reddy RB, Khora SS, Suresh A (2019) Molecular prognosticators in clinically and pathologically distinct cohorts of head and neck squamous cell carcinoma—A meta-analysis approach. PLoS ONE 14(7): e0218989.
- Scotto, L., Narayan, G., Nandula, S.V. et al. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer 7, 58 (2008).
- Google Patents