Alpha solenoid
An alpha solenoid (sometimes also known as an alpha horseshoe or as stacked pairs of alpha helices, abbreviated SPAH) is a protein fold composed of repeating alpha helix subunits, commonly helix-turn-helix motifs, arranged in antiparallel fashion to form a superhelix.[2] Alpha solenoids are known for their flexibility and plasticity.[3] Like beta propellers, alpha solenoids are a form of solenoid protein domain commonly found in the proteins comprising the nuclear pore complex.[4] They are also common in membrane coat proteins known as coatomers, such as clathrin, and in regulatory proteins that form extensive protein-protein interactions with their binding partners.[2][4] Examples of alpha solenoid structures binding RNA and lipids have also been described.[2]
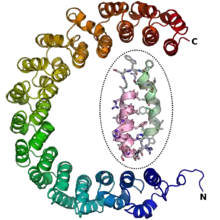
Terminology and classification
The term "alpha solenoid" has been used somewhat inconsistently in the literature.[4] As originally defined, alpha solenoids were composed of helix-turn-helix motifs that stacked into an open superhelix.[5] However, protein structural classification systems have used varying terminology; the Structural Classification of Proteins (SCOP) database describes these proteins using the term "alpha alpha superhelix". The CATH database uses the term "alpha horseshoe" [6] for these proteins, and uses "alpha solenoid" for a somewhat different and more compact structure exemplified by the peridinin-chlorophyll binding protein.[4]
Structure
Alpha solenoid proteins are composed of repeating structural units containing at least two alpha helices arranged in an antiparallel orientation. Often the repeating unit is a helix-turn-helix motif, but it can be more elaborate, as in variants with an additional helix in the turn segment.[2] Alpha solenoids can be formed by several different types of helical tandem repeats, including HEAT repeats, Armadillo repeats, tetratricopeptide (TPR) repeats, leucine-rich repeats, and ankyrin repeats.[2][4][5]
Alpha solenoids have unusual elasticity and flexibility relative to globular proteins.[2][3] They are sometimes considered to occupy an intermediate position between globular proteins and fibrous structural proteins, distinct from the latter in part due to the alpha solenoids' lack of need for intermolecular interactions to maintain their structure.[5] The extent of the curvature of an alpha solenoid superhelix varies considerably among the class, resulting in the ability of these proteins to form large, extended protein-protein interaction surfaces or to form deep concave areas for binding globular proteins.[2]
Because they are composed of repeating relatively short subunits, alpha solenoids can acquire additional subunits relatively easily, resulting in new interaction surface properties.[2] As a result, known alpha solenoid proteins vary substantially in length.[4]
Function
Nuclear pore complex components
Alpha solenoids feature prominently in the proteins making up the nuclear pore complex (NPC); alpha solenoid and beta propeller domains together account for up to half of the core NPC scaffold by mass.[4] A large number of the conserved nucleoporin proteins forming the NPC are either alpha solenoid proteins or consist of a beta propeller domain at the N-terminus and an alpha solenoid at the C-terminus.[7][8] This latter domain architecture also occurs in clathrin and Sec31, and was thought to be unique to eukaryotes,[7][9] though a few examples have been reported in planctomycetes.[10]
Vesicle coat proteins

Vesicle coat proteins frequently contain alpha solenoids and share common domain architecture with some NPC proteins.[7] Three major coat complexes involved in distinct cellular pathways all contain alpha solenoid proteins: the clathrin/adaptin complex, which buds vesicles from the plasma membrane and is involved in endocytosis; the COPI complex, which buds vesicles from the Golgi apparatus and is associated with retrograde transport; and the COPII complex, which buds vesicles from the endoplasmic reticulum and is associated with anterograde transport.[12]
Transport proteins
Due to their propensity for forming large interaction surfaces well-suited to protein-protein interactions, and their flexible surfaces permitting binding of various cargo molecules, alpha solenoid proteins commonly function as transport proteins, particularly in transport between the nucleus and the cytoplasm.[2] For example, the beta-karyopherin superfamily consists of alpha solenoid proteins formed from HEAT repeats; importin beta is a member of this family, and its adaptor protein importin alpha is an alpha solenoid formed from Armadillo repeats.[13] Transporters of other molecules, such as RNA, can also be of alpha solenoid architecture, as in exportin-5[14] or pentatricopeptide-repeat-containing RNA-binding proteins, which are particularly common in plants.[15][16]
Regulatory proteins
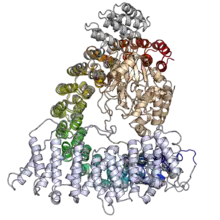
The protein-protein interaction capacity of alpha solenoid proteins also makes them well suited to function as regulatory proteins. For example, regulatory subunit A (also known as PR65) of protein phosphatase 2A is a HEAT-repeat alpha solenoid whose conformational flexibility regulates access to the enzyme binding site.[18][1]
Taxonomic distribution
Alpha solenoid proteins are found in all domains of life; however, their frequencies in different proteomes vary significantly. They are rare in viruses and bacteria, somewhat more common in archaea, and quite common in eukaryotes. Many of the eukaryotic alpha solenoid proteins have detectable homologs only in other eukaryotes and are often restricted even further, to the chordates. Prokaryotic alpha solenoid proteins are concentrated in particular taxa, notably the cyanobacteria and planctomycetes, which have unusually complex intracellular compartmentalization relative to most prokaryotes.[2]
Evolution
Evolutionary relationships between different alpha solenoid proteins are difficult to trace due to the low sequence homology of the repeats. Convergent evolution of similar protein structures from ancestrally unrelated proteins is thought to be significant in the evolutionary history of this fold class.[2]
Nuclear pore complexes and vesicle transport
The nuclear pore complex is an extremely large protein complex that mediates transit into and out of the cell nucleus. Homologous structures from which the NPC might have evolved have not been detected in prokaryotic transmembrane transport proteins; however, it has been suggested that the NPC components show distinct homology to vesicle coat proteins found in clathrin/adaptin, COPI, and COPII complexes. Most distinctively, a shared domain architecture consisting of an N-terminal beta propeller and a C-terminal alpha solenoid has been detected in both NPC and coat proteins, suggesting a possible common origin.[7][8] An ancestral "protocoatomer" that diversified to acquire derived characteristics of all four modern complexes has been proposed.[4][19][20][21]
Examination of the genome of Lokiarchaeum, thought to be among the closest archaeal relatives to eukaryotes, did not reveal any examples of the beta propeller/alpha solenoid domain architecture, although homologs of other proteins involved in eukaryotic membrane trafficking were identified. However, it is unclear whether this observation means that the propeller/solenoid architecture evolved later or was lost from modern lokiarchaea.[22]
Membrane coat proteins in prokaryotes
A survey of the sequenced genomes of complex prokaryotes from the PVC superphylum (Planctomycetes-Verrucomicrobia-Chlamydiae) identified examples of proteins with homology to eukaryotic membrane trafficking proteins, including examples of the distinctive beta-propeller/alpha-solenoid domain architecture previously believed to be unique to eukaryotes.[10] The PVC superphylum is known for containing bacteria with unusually complex membrane morphology, and this discovery has been cited as evidence in favor of these organisms' status as an intermediate form between prokaryotes and eukaryotes. The planctomycete Gemmata obscuriglobus has exceptionally complex membrane architecture and has been a source of controversy in the literature regarding the possibility that it has a membrane-bound "nucleoid" compartment enclosing its DNA.[23][24][25][26][27][28] The identification of proteins with sequence similarities to HEAT repeats in the G. obscuriglobus proteome has been interpreted as support for the membrane-bound nucleoid hypothesis;[29] however, this has been disputed.[24]
Bioinformatics
Low sequence similarity among alpha solenoid proteins of similar structure has impeded their identification using bioinformatics methods, since the repeats are often not well defined in sequence. A large number of different computational methods have been developed to identify candidate alpha solenoid proteins based on their amino acid sequence.[2][30][31]
External links
- RepeatsDB α-solenoid class
References
- Cho, Uhn Soo; Xu, Wenqing (1 November 2006). "Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme". Nature. 445 (7123): 53–57. doi:10.1038/nature05351. PMID 17086192. S2CID 4408160.
- Fournier, David; Palidwor, Gareth A.; Shcherbinin, Sergey; Szengel, Angelika; Schaefer, Martin H.; Perez-Iratxeta, Carol; Andrade-Navarro, Miguel A.; E. Tosatto, Silvio C. (21 November 2013). "Functional and Genomic Analyses of Alpha-Solenoid Proteins". PLOS ONE. 8 (11): e79894. Bibcode:2013PLoSO...879894F. doi:10.1371/journal.pone.0079894. PMC 3837014. PMID 24278209.
- Kappel, Christian; Zachariae, Ulrich; Dölker, Nicole; Grubmüller, Helmut (September 2010). "An Unusual Hydrophobic Core Confers Extreme Flexibility to HEAT Repeat Proteins". Biophysical Journal. 99 (5): 1596–1603. Bibcode:2010BpJ....99.1596K. doi:10.1016/j.bpj.2010.06.032. PMC 2931736. PMID 20816072.
- Field, Mark C.; Sali, Andrej; Rout, Michael P. (13 June 2011). "On a bender—BARs, ESCRTs, COPs, and finally getting your coat". The Journal of Cell Biology. 193 (6): 963–972. doi:10.1083/jcb.201102042. PMC 3115789. PMID 21670211.
- Kobe, Bostjan; Kajava, Andrey V (October 2000). "When protein folding is simplified to protein coiling: the continuum of solenoid protein structures". Trends in Biochemical Sciences. 25 (10): 509–515. doi:10.1016/S0968-0004(00)01667-4. PMID 11050437.
- "CATH Topology "Alpha Horseshoe"".
- Alber, Frank; Dokudovskaya, Svetlana; Veenhoff, Liesbeth M.; Zhang, Wenzhu; Kipper, Julia; Devos, Damien; Suprapto, Adisetyantari; Karni-Schmidt, Orit; Williams, Rosemary; Chait, Brian T.; Sali, Andrej; Rout, Michael P. (29 November 2007). "The molecular architecture of the nuclear pore complex". Nature. 450 (7170): 695–701. Bibcode:2007Natur.450..695A. doi:10.1038/nature06405. PMID 18046406. S2CID 4431057.
- Devos, Damien; Dokudovskaya, Svetlana; Alber, Frank; Williams, Rosemary; Chait, Brian T; Sali, Andrej; Rout, Michael P; Greg Petsko (2 November 2004). "Components of Coated Vesicles and Nuclear Pore Complexes Share a Common Molecular Architecture". PLOS Biology. 2 (12): e380. doi:10.1371/journal.pbio.0020380. PMC 524472. PMID 15523559.
- Antonin, Wolfram; Mattaj, Iain W. (January 2005). "Nuclear pore complexes: Round the bend?". Nature Cell Biology. 7 (1): 10–12. doi:10.1038/ncb0105-10. PMID 15632943. S2CID 9909704.
- Santarella-Mellwig, Rachel; Franke, Josef; Jaedicke, Andreas; Gorjanacz, Matyas; Bauer, Ulrike; Budd, Aidan; Mattaj, Iain W.; Devos, Damien P.; Schmid, Sandra L. (19 January 2010). "The Compartmentalized Bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae Superphylum Have Membrane Coat-Like Proteins". PLOS Biology. 8 (1): e1000281. doi:10.1371/journal.pbio.1000281. PMC 2799638. PMID 20087413.
- Ybe, Joel A.; Brodsky, Frances M.; Hofmann, Kay; Lin, Kai; Liu, Shu-Hui; Chen, Lin; Earnest, Thomas N.; Fletterick, Robert J.; Hwang, Peter K. (27 May 1999). "Clathrin self-assembly is mediated by a tandemly repeated superhelix". Nature. 399 (6734): 371–375. Bibcode:1999Natur.399..371Y. doi:10.1038/20708. PMID 10360576. S2CID 4406014.
- Lee, Changwook; Goldberg, Jonathan (July 2010). "Structure of Coatomer Cage Proteins and the Relationship among COPI, COPII, and Clathrin Vesicle Coats". Cell. 142 (1): 123–132. doi:10.1016/j.cell.2010.05.030. PMC 2943847. PMID 20579721.
- Forwood, Jade K.; Lange, Allison; Zachariae, Ulrich; Marfori, Mary; Preast, Callie; Grubmüller, Helmut; Stewart, Murray; Corbett, Anita H.; Kobe, Bostjan (September 2010). "Quantitative Structural Analysis of Importin-β Flexibility: Paradigm for Solenoid Protein Structures". Structure. 18 (9): 1171–1183. doi:10.1016/j.str.2010.06.015. hdl:11858/00-001M-0000-0027-C07B-1. PMID 20826343.
- Katahira, Jun; Yoneda, Yoshihiro (November 2011). "Nucleocytoplasmic Transport of MicroRNAs and Related Small RNAs". Traffic. 12 (11): 1468–1474. doi:10.1111/j.1600-0854.2011.01211.x. PMID 21518166.
- Barkan, Alice; Rojas, Margarita; Fujii, Sota; Yap, Aaron; Chong, Yee Seng; Bond, Charles S.; Small, Ian; Voytas, Dan (16 August 2012). "A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins". PLOS Genetics. 8 (8): e1002910. doi:10.1371/journal.pgen.1002910. PMC 3420917. PMID 22916040.
- Barkan, Alice; Small, Ian (29 April 2014). "Pentatricopeptide Repeat Proteins in Plants". Annual Review of Plant Biology. 65 (1): 415–442. doi:10.1146/annurev-arplant-050213-040159. PMID 24471833.
- Groves, Matthew R.; Hanlon, Neil; Turowski, Patric; Hemmings, Brian A.; Barford, David (January 1999). "The Structure of the Protein Phosphatase 2A PR65/A Subunit Reveals the Conformation of Its 15 Tandemly Repeated HEAT Motifs". Cell. 96 (1): 99–110. doi:10.1016/S0092-8674(00)80963-0. PMID 9989501. S2CID 14465060.
- Grinthal, A.; Adamovic, I.; Weiner, B.; Karplus, M.; Kleckner, N. (25 January 2010). "PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis". Proceedings of the National Academy of Sciences. 107 (6): 2467–2472. Bibcode:2010PNAS..107.2467G. doi:10.1073/pnas.0914073107. PMC 2823866. PMID 20133745.
- Field, Mark C; Dacks, Joel B (February 2009). "First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes". Current Opinion in Cell Biology. 21 (1): 4–13. doi:10.1016/j.ceb.2008.12.004. PMID 19201590.
- Dacks, Joel B.; Field, Mark C.; Buick, Roger; Eme, Laura; Gribaldo, Simonetta; Roger, Andrew J.; Brochier-Armanet, Céline; Devos, Damien P. (26 September 2016). "The changing view of eukaryogenesis – fossils, cells, lineages and how they all come together". Journal of Cell Science. 129 (20): 3695–3703. doi:10.1242/jcs.178566. PMID 27672020.
- Promponas, Vasilis J.; Katsani, Katerina R.; Blencowe, Benjamin J.; Ouzounis, Christos A. (2 March 2016). "Sequence evidence for common ancestry of eukaryotic endomembrane coatomers". Scientific Reports. 6: 22311. Bibcode:2016NatSR...622311P. doi:10.1038/srep22311. PMC 4773986. PMID 26931514.
- Klinger, Christen M.; Spang, Anja; Dacks, Joel B.; Ettema, Thijs J.G. (June 2016). "Tracing the Archaeal Origins of Eukaryotic Membrane-Trafficking System Building Blocks". Molecular Biology and Evolution. 33 (6): 1528–1541. doi:10.1093/molbev/msw034. PMID 26893300.
- Fuerst, John A. (2010). "Beyond Prokaryotes and Eukaryotes : Planctomycetes and Cell Organization". Nature Education. 3 (9): 44.
- McInerney, JO; Martin, WF; Koonin, EV; Allen, JF; Galperin, MY; Lane, N; Archibald, JM; Embley, TM (November 2011). "Planctomycetes and eukaryotes: a case of analogy not homology". BioEssays. 33 (11): 810–7. doi:10.1002/bies.201100045. PMC 3795523. PMID 21858844.
- Fuerst, JA (October 2013). "The PVC superphylum: exceptions to the bacterial definition?". Antonie van Leeuwenhoek. 104 (4): 451–66. doi:10.1007/s10482-013-9986-1. PMID 23912444. S2CID 14283647.
- Devos, Damien P. (September 2013). "Gemmata obscuriglobus". Current Biology. 23 (17): R705–R707. doi:10.1016/j.cub.2013.07.013. PMID 24028944.
- Devos, DP (February 2014). "Re-interpretation of the evidence for the PVC cell plan supports a Gram-negative origin". Antonie van Leeuwenhoek. 105 (2): 271–4. doi:10.1007/s10482-013-0087-y. hdl:10261/129395. PMID 24292377. S2CID 16557669.
- Devos, Damien P. (January 2014). "PVC bacteria: variation of, but not exception to, the Gram-negative cell plan". Trends in Microbiology. 22 (1): 14–20. doi:10.1016/j.tim.2013.10.008. hdl:10261/129431. PMID 24286661.
- Fuerst, John A.; Sagulenko, Evgeny (August 2014). "Towards understanding the molecular mechanism of the endocytosis-like process in the bacterium Gemmata obscuriglobus". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1843 (8): 1732–1738. doi:10.1016/j.bbamcr.2013.10.002. PMID 24144586.
- Di Domenico, Tomás; Potenza, Emilio; Walsh, Ian; Gonzalo Parra, R.; Giollo, Manuel; Minervini, Giovanni; Piovesan, Damiano; Ihsan, Awais; Ferrari, Carlo; Kajava, Andrey V.; Tosatto, Silvio C.E. (January 2014). "RepeatsDB: a database of tandem repeat protein structures". Nucleic Acids Research. 42 (D1): D352–D357. doi:10.1093/nar/gkt1175. PMC 3964956. PMID 24311564.
- Pellegrini, Marco (24 September 2015). "Tandem Repeats in Proteins: Prediction Algorithms and Biological Role". Frontiers in Bioengineering and Biotechnology. 3: 143. doi:10.3389/fbioe.2015.00143. PMC 4585158. PMID 26442257.
