Bacterial small RNA
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops.[1][2] Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq[3] in a number of bacterial species including Escherichia coli,[4][5][6] the model pathogen Salmonella,[7] the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti,[8] marine cyanobacteria,[9] Francisella tularensis (the causative agent of tularaemia),[10] Streptococcus pyogenes[11], the pathogen Staphylococcus aureus[12], and the plant pathogen Xanthomonas oryzae pathovar oryzae.[13] Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.[7][12]
Origin
In the 1960s, the abbreviation sRNA was used to refer to "soluble RNA," which is now known as transfer RNA or tRNA (for an example of the abbreviation used in this sense, see[14]). It is now known that most bacterial sRNAs are encoded by free-standing genes located in the intergenic regions (IGR) between two known genes.[3][6] However, a class of sRNAs are shown to be derived from the 3'-UTR of mRNAs by independent transcription or nucleolytic cleavage.[15]
The first bacterial sRNA was discovered and characterized in 1984. MicF in E. coli was found to regulate the expression of a key structural gene that makes up the outer membrane of the E. coli cell.[16] Shortly after, the Staphylococcus aureus sRNA RNAIII was found to act as a global regulator of S. aureus virulence and toxin secretion.[16] Since these initial discoveries, over six thousand bacterial sRNAs have been identified, largely through RNA-sequencing experiments.[17]
Techniques
Several laboratory and bioinformatic techniques can be used to identify and characterize sRNA transcripts.[3]
- RNA-sequencing, or RNA-seq, is used to analyze expression levels of all transcripts in a genome, including sRNAs.[18]
- Microarrays use complementary DNA probes to bind to possible sRNA loci in intergenic regions.[3]
- Northern blotting can reveal possible sRNA transcript size and expression levels by running a mixed RNA sample on an agarose gel and probing for a desired sRNA.[3]
- Target prediction software can predict possible interactions between sRNAs and mRNA by finding regions of complementarity within sRNA and mRNA target sequences.[19]
- RNase crosslinking can experimentally validate sRNA and mRNA interactions by crosslinking a sRNA and its target with UV light, along with RNase enzymes that are also usually involved in the interaction. The sRNA:mRNA hybrid can then be isolated and analyzed.[20]
Function
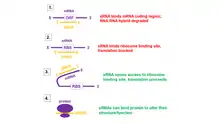
Bacterial sRNAs have a wide variety of regulatory mechanisms. Generally, sRNAs can bind to protein targets and modify the function of the bound protein.[21] Alternately, sRNAs may interact with mRNA targets and regulate gene expression by binding to complementary mRNA and blocking translation, or by unmasking or blocking the ribosome-binding site.[21]
sRNAs that interact with mRNA can also be categorized as cis- or trans-acting. Cis-acting sRNAs interact with genes encoded on the same genetic locus as the sRNA.[22] Some cis-acting sRNAs act as riboswitches, which have receptors for specific environmental or metabolic signals and activate or repress genes based on these signals.[16] Conversely, trans-encoded sRNAs interact with genes on separate loci.[1]
House-keeping
Amongst the targets of sRNAs are a number of house-keeping genes. The 6S RNA binds to RNA polymerase and regulates transcription, tmRNA has functions in protein synthesis, including the recycling of stalled ribosomes, 4.5S RNA regulates signal recognition particle (SRP), which is required for the secretion of proteins and RNase P is involved in maturing tRNAs.[23][24]
Stress response
Many sRNAs are involved in stress response regulation.[25] They are expressed under stress conditions such as cold shock, iron depletion, onset of the SOS response and sugar stress.[24] The small RNA nitrogen stress-induced RNA 1 (NsiR1) is produced by Cyanobacteria under conditions of nitrogen deprivation.[26] Cyanobacteria NisR8 and NsiR9 sRNAs could be related to the differentiation of nitrogen-fixing cells (heterocysts).[27]
Regulation of RpoS
The RpoS gene in E. coli encodes sigma 38, a sigma factor which regulates stress response and acts as a transcriptional regulator for many genes involved in cell adaptation. At least three sRNAs, DsrA, RprA and OxyS, regulate the translation of RpoS. DsrA and RprA both activate RpoS translation by base pairing to a region in the leader sequence of the RpoS mRNA and disrupting formation of a hairpin which frees up the ribosome loading site. OxyS inhibits RpoS translation. DsrA levels are increased in response to low temperatures and osmotic stress, and RprA levels are increased in response to osmotic stress and cell-surface stress, therefore increasing RpoS levels in response to these conditions. Levels of OxyS are increased in response to oxidative stress, therefore inhibiting RpoS under these conditions.[24][28][29]
Regulation of outer membrane proteins
The outer membrane of gram negative bacteria acts as a barrier to prevent the entry of toxins into the bacterial cell, and plays a role in the survival of bacterial cells in diverse environments. Outer membrane proteins (OMPs) include porins and adhesins. Numerous sRNAs regulate the expression of OMPs. The porins OmpC and OmpF are responsible for the transport of metabolites and toxins. The expression of OmpC and OmpF is regulated by the sRNAs MicC and MicF in response to stress conditions.[30][31][32] The outer membrane protein OmpA anchors the outer membrane to the murein layer of the periplasmic space. Its expression is downregulated in the stationary phase of cell-growth. In E. coli the sRNA MicA depletes OmpA levels, in Vibrio cholerae the sRNA VrrA represses synthesis of OmpA in response to stress.[30][33]
Virulence
In some bacteria sRNAs regulate virulence genes. In Salmonella, the pathogenicity island encoded InvR RNA represses synthesis of the major outer membrane protein OmpD; another co-activated DapZ sRNA from 3'-UTR represses abundant membrane Opp/Dpp transporters of oligopeptides;[15] and SgrS sRNA regulates the expression of the secreted effector protein SopD.[7] In Staphylococcus aureus, RNAIII regulates a number of genes involved in toxin and enzyme production and cell-surface proteins.[24] The FasX sRNA is the only well-characterized regulatory RNA known to control the regulation of several virulence factors in Streptococcus pyogenes, including both cell-surface associated adhesion proteins as well as secreted factors.[34][35][36][37]
Quorum sensing
In Vibrio species, the Qrr sRNAs and the chaperone protein Hfq are involved in the regulation of quorum sensing. Qrr sRNAs regulate the expression of several mRNAs including the quorum-sensing master regulators LuxR and HapR.[38][39]
Biofilm Formation
Biofilm is a type of bacterial growth pattern where multiple layers of bacterial cells adhere to a host surface. This mode of growth is often found in pathogenic bacteria, including Pseudomonas aeruginosa, which can form persistent biofilm within the respiratory tract and cause chronic infection.[40] The P. aeruginosa sRNA SbrA was found to be necessary for full biofilm formation and pathogenicity.[40] A mutant P. aeruginosa strain with SbrA deleted formed a 66% smaller biofilm and its ability to infect a nematode model was reduced by nearly half when compared to wildtype P. aeruginosa.[40]
Antibiotic Resistance
Several bacterial sRNAs are involved in the regulation of genes that confer antibiotic resistance.[41] For example, the sRNA DsrA regulates a drug efflux pump in E. coli, which is a system that mechanically pumps antibiotic out of bacterial cells.[41] E. coli MicF also contributes to antibiotic resistance of cephalosporins, as it regulates membrane proteins involved in uptake of these class of antibiotics.[41]
Target prediction
In order to understand an sRNA's function one primarily needs to describe its targets. Here, target predictions represent a fast and free method for initial characterization of putative targets, given that the sRNA actually exerts its function via direct base pairing with a target RNA. Examples are CopraRNA,[42][43] IntaRNA,[43][44][45] TargetRNA[19] and RNApredator.[46] It has been shown that target prediction for enterobacterial sRNAs can benefit from transcriptome wide Hfq-binding maps.[47]
Databases
- BSRD (kwanlab.bio.cuhk.edu.hk/BSRD) is a repository for published sRNA sequences with multiple annotations and expression profiles.[17]
- SRD (srd.genouest.org/) is a database of Staphylococcus aureus sRNAs with sequences, predicted structures, and genome start and end sites.[48]
- sRNAdb (http://srnadb.fb11.uni-giessen.de/sRNAdb) is a databse of sRNAs from Gram-positive bacterial species with sequence annotation.[49]
See also
- 5 prime ureB sRNA
- Aar small RNA, an sRNA produced by species of Acinetobacter
- Bacillus subtilis BSR sRNAs
- Escherichia coli sRNA
- Mycobacterium tuberculosis sRNA
- Non-coding RNA
- Xanthomonas sRNA
- Brucella sRNA
- Anti small RNA
- Riboswitches
- MicF, the first characterized chromosomal sRNA
- RNAIII, the first characterized bacterial sRNA found to influence virulence
References
- Vogel J, Wagner EG (June 2007). "Target identification of small noncoding RNAs in bacteria". Curr. Opin. Microbiol. 10 (3): 262–270. doi:10.1016/j.mib.2007.06.001. PMID 17574901.
- Viegas SC, Arraiano CM (2008). "Regulating the regulators: How ribonucleases dictate the rules in the control of small non-coding RNAs". RNA Biol. 5 (4): 230–243. doi:10.4161/rna.6915. PMID 18981732.
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S (July 2001). "Identification of novel small RNAs using comparative genomics and microarrays". Genes Dev. 15 (13): 1637–1651. doi:10.1101/gad.901001. PMC 312727. PMID 11445539.
- Hershberg R, Altuvia S, Margalit H (April 2003). "A survey of small RNA-encoding genes in Escherichia coli". Nucleic Acids Res. 31 (7): 1813–1820. doi:10.1093/nar/gkg297. PMC 152812. PMID 12654996.
- Rivas E, Klein RJ, Jones TA, Eddy SR (September 2001). "Computational identification of noncoding RNAs in E. coli by comparative genomics". Curr. Biol. 11 (17): 1369–1373. doi:10.1016/S0960-9822(01)00401-8. PMID 11553332.
- Argaman L, Hershberg R, Vogel J, et al. (June 2001). "Novel small RNA-encoding genes in the intergenic regions of Escherichia coli". Curr. Biol. 11 (12): 941–950. doi:10.1016/S0960-9822(01)00270-6. PMID 11448770.
- Vogel J (January 2009). "A rough guide to the non-coding RNA world of Salmonella". Mol. Microbiol. 71 (1): 1–11. doi:10.1111/j.1365-2958.2008.06505.x. PMID 19007416.
- Schlüter JP, Reinkensmeier J, Daschkey S, et al. (2010). "A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti". BMC Genomics. 11: 245. doi:10.1186/1471-2164-11-245. PMC 2873474. PMID 20398411.
- Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR (2005). "Identification of cyanobacterial non-coding RNAs by comparative genome analysis". Genome Biol. 6 (9): R73. doi:10.1186/gb-2005-6-9-r73. PMC 1242208. PMID 16168080.
- Postic G, Frapy E, Dupuis M, et al. (2010). "Identification of small RNAs in Francisella tularensis". BMC Genomics. 11: 625. doi:10.1186/1471-2164-11-625. PMC 3091763. PMID 21067590.
- Tesorero, Rafael A.; Yu, Ning; Wright, Jordan O.; Svencionis, Juan P.; Cheng, Qiang; Kim, Jeong-Ho; Cho, Kyu Hong (2013-01-01). "Novel regulatory small RNAs in Streptococcus pyogenes". PLOS One. 8 (6): e64021. doi:10.1371/journal.pone.0064021. ISSN 1932-6203. PMC 3675131. PMID 23762235.
- Felden, Brice; Vandenesch, François; Bouloc, Philippe; Romby, Pascale (2011-03-10). "The Staphylococcus aureus RNome and Its Commitment to Virulence". PLoS Pathogens. 7 (3): e1002006. doi:10.1371/journal.ppat.1002006. ISSN 1553-7366. PMC 3053349. PMID 21423670.
- Liang H, Zhao YT, Zhang JQ, Wang XJ, Fang RX, Jia YT (2011). "Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae". BMC Genomics. 12: 87. doi:10.1186/1471-2164-12-87. PMC 3039613. PMID 21276262.
- Crick F (1966). "Codon–anticodon pairing: the wobble hypothesis" (PDF). J Mol Biol. 19 (2): 548–555. doi:10.1016/S0022-2836(66)80022-0. PMID 5969078.
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J (October 2012). "An atlas of Hfq-bound transcripts reveals 3' UTRs as a genomic reservoir of regulatory small RNAs". EMBO J. 31 (20): 4005–4019. doi:10.1038/emboj.2012.229. PMC 3474919. PMID 22922465.
- Svensson, Sarah L.; Sharma, Cynthia M. (June 2016). "Small RNAs in Bacterial Virulence and Communication". Microbiology Spectrum. 4 (3): 169–212. doi:10.1128/microbiolspec.VMBF-0028-2015. ISSN 2165-0497. PMID 27337442.
- Li, L; Kwan, HS (January 2013). "BSRD: a repository for bacterial small regulatory RNA". Nucleic Acids Research. 41 (Database issue): D233-8. doi:10.1093/nar/gks1264. PMC 3531160. PMID 23203879.
- Kanniappan, Priyatharisni; Ahmed, Siti Aminah; Rajasekaram, Ganeswrie; Marimuthu, Citartan; Ch'ng, Ewe Seng; Lee, Li Pin; Raabe, Carsten A.; Rozhdestvensky, Timofey S.; Tang, Thean Hock (October 2017). "RNomic identification and evaluation of npcTB_6715, a non-protein-coding RNA gene as a potential biomarker for the detection of Mycobacterium tuberculosis". Journal of Cellular and Molecular Medicine. 21 (10): 2276–2283. doi:10.1111/jcmm.13148. ISSN 1582-4934. PMC 5618688. PMID 28756649.
- Tjaden B, Goodwin SS, Opdyke JA, et al. (2006). "Target prediction for small, noncoding RNAs in bacteria". Nucleic Acids Res. 34 (9): 2791–2802. doi:10.1093/nar/gkl356. PMC 1464411. PMID 16717284.
- Waters, Shafagh A.; McAteer, Sean P.; Kudla, Grzegorz; Pang, Ignatius; Deshpande, Nandan P.; Amos, Timothy G.; Leong, Kai Wen; Wilkins, Marc R.; Strugnell, Richard (2017-02-01). "Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E". The EMBO Journal. 36 (3): 374–387. doi:10.15252/embj.201694639. ISSN 1460-2075. PMC 5286369. PMID 27836995.
- Waters, Lauren S.; Storz, Gisela (2009-02-20). "Regulatory RNAs in bacteria". Cell. 136 (4): 615–628. doi:10.1016/j.cell.2009.01.043. ISSN 1097-4172. PMC 3132550. PMID 19239884.
- Guillet, Julien; Hallier, Marc; Felden, Brice (2013). "Emerging functions for the Staphylococcus aureus RNome". PLoS Pathogens. 9 (12): e1003767. doi:10.1371/journal.ppat.1003767. ISSN 1553-7374. PMC 3861533. PMID 24348246.
- Wassarman KM (April 2007). "6S RNA: a small RNA regulator of transcription". Curr. Opin. Microbiol. 10 (2): 164–168. doi:10.1016/j.mib.2007.03.008. PMID 17383220.
- Christian Hammann; Nellen, Wolfgang (2005). Small RNAs:: Analysis and Regulatory Functions (Nucleic Acids and Molecular Biology). Berlin: Springer. ISBN 978-3-540-28129-0.
- Caswell CC, Oglesby-Sherrouse AG, Murphy ER (October 2014). "Sibling rivalry: related bacterial small RNAs and their redundant and non-redundant roles". Front Cell Infect Microbiol. 4: 151. doi:10.3389/fcimb.2014.00151. PMC 4211561. PMID 25389522.
- Ionescu, D; Voss, B; Oren, A; Hess, WR; Muro-Pastor, AM (Apr 30, 2010). "Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria". Journal of Molecular Biology. 398 (2): 177–188. doi:10.1016/j.jmb.2010.03.010. hdl:10261/112252. PMID 20227418.
- Brenes-Álvarez, Manuel; Olmedo-Verd, Elvira; Vioque, Agustín; Muro-Pastor, Alicia M. (2016-01-01). "Identification of Conserved and Potentially Regulatory Small RNAs in Heterocystous Cyanobacteria". Frontiers in Microbiology. 7: 48. doi:10.3389/fmicb.2016.00048. ISSN 1664-302X. PMC 4734099. PMID 26870012.
- Repoila F, Majdalani N, Gottesman S (May 2003). "Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm". Mol. Microbiol. 48 (4): 855–861. doi:10.1046/j.1365-2958.2003.03454.x. PMID 12753181.
- Benjamin JA, Desnoyers G, Morissette A, Salvail H, Massé E (March 2010). "Dealing with oxidative stress and iron starvation in microorganisms: an overview". Can. J. Physiol. Pharmacol. 88 (3): 264–272. doi:10.1139/y10-014. PMID 20393591.
- Vogel J, Papenfort K (December 2006). "Small non-coding RNAs and the bacterial outer membrane". Curr. Opin. Microbiol. 9 (6): 605–611. doi:10.1016/j.mib.2006.10.006. PMID 17055775.
- Delihas N, Forst S (October 2001). "MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors". J. Mol. Biol. 313 (1): 1–12. doi:10.1006/jmbi.2001.5029. PMID 11601842.
- Chen S, Zhang A, Blyn LB, Storz G (October 2004). "MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli". J. Bacteriol. 186 (20): 6689–6697. doi:10.1128/JB.186.20.6689-6697.2004. PMC 522180. PMID 15466019.
- Song T, Wai SN (July 2009). "A novel sRNA that modulates virulence and environmental fitness of Vibrio cholerae". RNA Biol. 6 (3): 254–258. doi:10.4161/rna.6.3.8371. PMID 19411843.
- Ramirez-Peña, E; Treviño, J; Liu, Z; Perez, N; Sumby, P (December 2010). "The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript". Molecular Microbiology. 78 (6): 1332–1347. doi:10.1111/j.1365-2958.2010.07427.x. PMC 3071709. PMID 21143309.
- Liu, Z; Treviño, J; Ramirez-Peña, E; Sumby, P (October 2012). "The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus". Molecular Microbiology. 86 (1): 140–154. doi:10.1111/j.1365-2958.2012.08178.x. PMC 3456998. PMID 22882718.
- Danger, JL; Cao, TN; Cao, TH; Sarkar, P; Treviño, J; Pflughoeft, KJ; Sumby, P (April 2015). "The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets". Molecular Microbiology. 96 (2): 249–262. doi:10.1111/mmi.12935. PMC 4390479. PMID 25586884.
- Danger, JL; Makthal, N; Kumaraswami, M; Sumby, P (1 December 2015). "The FasX Small Regulatory RNA Negatively Regulates the Expression of Two Fibronectin-Binding Proteins in Group A Streptococcus". Journal of Bacteriology. 197 (23): 3720–3730. doi:10.1128/jb.00530-15. PMC 4626899. PMID 26391206.
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL (July 2004). "The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae". Cell. 118 (1): 69–82. doi:10.1016/j.cell.2004.06.009. PMID 15242645.
- Bardill JP, Zhao X, Hammer BK (April 2011). "The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base-pairing interactions". Mol Microbiol. 80 (5): 1381–1394. doi:10.1111/j.1365-2958.2011.07655.x. PMID 21453446.
- Taylor, Patrick K.; Van Kessel, Antonius T. M.; Colavita, Antonio; Hancock, Robert E. W.; Mah, Thien-Fah (2017). "A novel small RNA is important for biofilm formation and pathogenicity in Pseudomonas aeruginosa". PLOS One. 12 (8): e0182582. doi:10.1371/journal.pone.0182582. ISSN 1932-6203. PMC 5542712. PMID 28771593.
- Dersch, Petra; Khan, Muna A.; Mühlen, Sabrina; Görke, Boris (2017). "Roles of Regulatory RNAs for Antibiotic Resistance in Bacteria and Their Potential Value as Novel Drug Targets". Frontiers in Microbiology. 8: 803. doi:10.3389/fmicb.2017.00803. ISSN 1664-302X. PMC 5418344. PMID 28529506.
- Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J (2013). "Comparative genomics boosts target prediction for bacterial small RNAs". Proc Natl Acad Sci U S A. 110 (37): E3487–E3496. doi:10.1073/pnas.1303248110. PMC 3773804. PMID 23980183.
- Wright PR, Georg J, Mann M, Sorescu DA, Richter AS, Lott S, Kleinkauf R, Hess WR, Backofen R (2014). "CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains". Nucleic Acids Res. 42 (Web Server): W119–23. CiteSeerX 10.1.1.641.51. doi:10.1093/nar/gku359. PMC 4086077. PMID 24838564.
- Busch A, Richter AS, Backofen R (2008). "IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions". Bioinformatics. 24 (24): 2849–2856. doi:10.1093/bioinformatics/btn544. PMC 2639303. PMID 18940824.
- Mann M, Wright PR, Backofen R (2017). "IntaRNA 2.0: enhanced and customizable prediction of RNA–RNA interactions". Nucleic Acids Res. 45 (Web Server): W435–439. doi:10.1093/nar/gkx279. PMC 5570192. PMID 28472523.
- Eggenhofer F, Tafer H, Stadler PF, Hofacker IL (2011). "RNApredator: fast accessibility-based prediction of sRNA targets". Nucleic Acids Res. 39 (Web Server): W149–154. doi:10.1093/nar/gkr467. PMC 3125805. PMID 21672960.
- Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J (2016). "Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo". EMBO J. 35: 991–1011. doi:10.15252/embj.201593360. PMC 5207318. PMID 27044921.
- Sassi, Mohamed; Augagneur, Yoann; Mauro, Tony; Ivain, Lorraine; Chabelskaya, Svetlana; Hallier, Marc; Sallou, Olivier; Felden, Brice (May 2015). "SRD: a Staphylococcus regulatory RNA database". RNA. 21 (5): 1005–1017. doi:10.1261/rna.049346.114. ISSN 1469-9001. PMC 4408781. PMID 25805861.
- Pischimarov, Jordan; Kuenne, Carsten; Billion, André; Hemberger, Jüergen; Cemič, Franz; Chakraborty, Trinad; Hain, Torsten (2012-08-10). "sRNAdb: a small non-coding RNA database for gram-positive bacteria". BMC Genomics. 13: 384. doi:10.1186/1471-2164-13-384. ISSN 1471-2164. PMC 3439263. PMID 22883983.