Vibrio cholerae
Vibrio cholerae is a Gram-negative, comma-shaped bacterium. The bacterium's natural habitat is brackish or saltwater where they attach themselves easily to the chitin-containing shells of crabs, shrimps, and other shellfish. Some strains of V. cholerae cause the disease cholera, which can be derived from the consumption of undercooked or raw marine life species. V. cholerae is a facultative anaerobe[1] and has a flagellum at one cell pole as well as pili. V. cholerae can undergo respiratory and fermentative metabolism. When ingested, V. cholerae can cause diarrhea and vomiting in a host within several hours to 2–3 days of ingestion. V. cholerae was first isolated as the cause of cholera in 1854 by Italian anatomist Filippo Pacini[2] and by the Catalan Joaquim Balcells i Pascual in the same year,[3][4] but their discovery was not widely known until Robert Koch, working independently 30 years later, publicized the knowledge and the means of fighting the disease.[5][6]
| Vibrio cholerae | |
|---|---|
 | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Proteobacteria |
| Class: | Gammaproteobacteria |
| Order: | Vibrionales |
| Family: | Vibrionaceae |
| Genus: | Vibrio |
| Species: | V. cholerae |
| Binomial name | |
| Vibrio cholerae Filippo Pacini & Robert Koch, 1854 | |
Characteristics
V. cholerae is a highly motile, comma shaped, halophilic, gram-negative rod. Initial isolates are slightly curved, whereas they can appear as straight rods upon laboratory culturing. The bacterium has a flagellum at one cell pole as well as pili. The Vibrios tolerate alkaline media that kill most intestinal commensals, but they are sensitive to acid. V. cholerae is a facultative anaerobe, and can undergo respiratory and fermentative metabolism.[1] It measures 0.3 micron in diameter and 1.3 micron in length[7] with average swimming velocity of around 75.4 +/- 9.4 microns/sec.[8]
Pathogenicity
V. cholerae pathogenicity genes code for proteins directly or indirectly involved in the virulence of the bacteria. To adapt the host intestinal environment and to avoid being attacked by bile acids and antimicrobial peptides, V. cholera used its outer membrane vesicles (OMVs). Upon entry, the bacteria sheds its OMVs, containing all the membrane modifications that make it vulnerable for the host attack.[9]
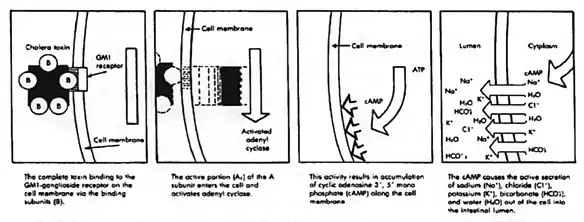
During infection, V. cholerae secretes cholera toxin (CT), a protein that causes profuse, watery diarrhea (known as "rice-water stool"). This cholera toxin contains 5 B subunits that plays a role in attaching to the intestinal epithelial cells and 1 A subunit that plays a role in toxin activity. Colonization of the small intestine also requires the toxin coregulated pilus (TCP), a thin, flexible, filamentous appendage on the surface of bacterial cells. Expression of both CT and TCP is mediated by two component systems (TCS), which typically consist of a membrane-bound histidine kinase and an intracellular response element.[10] TCS enable bacteria to respond to changing environments.[10] In V. cholerae several TCS have been identified to be important in colonization, biofilm production and virulence.[10] Recently, small RNAs (sRNA) have been identified as targets of V. cholerae TCS.[10][11][12] Here, sRNA molecules bind to mRNA to block translation or induce degradation of inhibitors of expression of virulence or colonization genes.[10][11] In V. cholerae the TCS EnvZ/OmpR alters gene expression via the sRNA coaR in response to changes in osmolarity and pH. An important target of coaR is tcpI, which negatively regulates expresion of the major subunit of theTCP encoding gene (tcpA). When tcpI is bound by coaR it is no longer able to repress expression tcpA, leading to an increased colonization ability.[10] Expression of coaR is upregulated by EnvZ/OmpR at a pH of 6,5, which is the normal pH of the intestinal lumen, but is low at higher pH values.[10] V. cholerae in the intestinal lumen utilizes the TCP to attach to the intestinal mucosa, not invading the mucosa.[10] After doing so it secretes cholerae toxin causing its symptoms. This then increases cyclic AMP or cAMP by binding (cholerae toxin) to adenylyl cyclase activating the GS pathway which leads to efflux of water and sodium into the intestinal lumen causing watery stools or rice watery stools. V. cholerae can cause syndromes ranging from asymptomatic to cholera gravis.[6] In endemic areas, 75% of cases are asymptomatic, 20% are mild to moderate, and 2-5% are severe forms such as cholera gravis.[6] Symptoms include abrupt onset of watery diarrhea (a grey and cloudy liquid), occasional vomiting, and abdominal cramps.[1][6] Dehydration ensues, with symptoms and signs such as thirst, dry mucous membranes, decreased skin turgor, sunken eyes, hypotension, weak or absent radial pulse, tachycardia, tachypnea, hoarse voice, oliguria, cramps, kidney failure, seizures, somnolence, coma, and death.[1] Death due to dehydration can occur in a few hours to days in untreated children. The disease is also particularly dangerous for pregnant women and their fetuses during late pregnancy, as it may cause premature labor and fetal death.[6][13][14] A study done by the Centers for Disease Control (CDC) in Haiti found that in pregnant women who contracted the disease, 16% of 900 women had fetal death. Risk factors for these deaths include: third trimester, younger maternal age, severe dehydration, and vomiting[15] Dehydration poses the biggest health risk to pregnant women in countries that there are high rates of cholera. In cases of cholera gravis involving severe dehydration, up to 60% of patients can die; however, less than 1% of cases treated with rehydration therapy are fatal. The disease typically lasts 4–6 days.[6][16] Worldwide, diarrhoeal disease, caused by cholera and many other pathogens, is the second-leading cause of death for children under the age of 5 and at least 120,000 deaths are estimated to be caused by cholera each year.[17][18] In 2002, the WHO deemed that the case fatality ratio for cholera was about 3.95%.[6]
Cholera illness and symptoms
.jpg.webp)
Cholera is an illness that derives from the bacteria, V. cholerae. This bacteria infects the intestine where it then causes diarrhea. This bacteria, V. cholerae can be spread by eating contaminated food or drinking contaminated water.[19] This illness is also spread through humans making skin contact with contaminated water from human feces. When it comes to symptoms, not everyone with Cholera will experience symptoms but it averages about 1 in 10 people with Cholera will experience symptoms. Some symptoms include: watery diarrhea, vomiting, rapid heart rate, loss of skin elasticity, low blood pressure, thirst, and muscle cramps.[19] This illness can get as serious as kidney failure and possible coma. If this illness is treated fast enough, the people infected can easily be cured and there is no chance of this illness reoccurring unless they are re-exposed to the bacteria.[19]
Disease occurrence
V. cholerae has an endemic or epidemic occurrence. In countries where the disease has been for the past three years and the cases confirmed are local (within the confines of the country) transmission is considered to be "endemic."[20]" Alternatively, an outbreak is declared when the occurrence of disease exceeds the normal occurrence for any given time or location.[21] Epidemics can last several days or over a span of years. Additionally, countries that have an occurrence of an epidemic can also be endemic.[21] The longest standing V. chloerae epidemic was recorded in Yemen. Yemen had two outbreaks, the first occurred between September 2016 and April 2017, and the second began later in April 2017 and recently was considered to be resolved in 2019.[22] The epidemic in Yemen took over 2,500 lives and impacted over 1 million people of Yemen[22] More outbreaks have occurred in Africa, the Americas, and Haiti.
Preventive measures
When visiting areas with epidemic cholera, the following precautions should be observed: drink and use bottled water; frequently wash hands with soap and safe water; use chemical toilets or bury feces if no restroom is available; do not defecate in any body of water and cook food thoroughly. Supplying proper, safe water is important.[23] A precaution to take is to properly sanitize.[24] Hand hygiene is an essential in areas where soap and water is not available. When there is no sanitation available for hand washing, scrub hands with ash or sand and rinse with clean water.[25] A single dose vaccine is available for those traveling to an area where cholera is common.
There is a V. cholerae vaccine available to prevent disease spread. The vaccine is known as the, "oral cholera vaccine" (OCV). There are three types of OCV available for prevention: Dukoral®, Shanchol™, and Euvichol-Plus®. All three OCVs require two doses to be fully effective. Countries who are endemic or have an epidemic status are eligible to receive the vaccine based on several criteria: Risk of cholera, Severity of cholera, WASH conditions and capacity to improve, Healthcare conditions and capacity to improve, Capacity to implement OCV campaigns, Capacity to conduct M&E activities, Commitment at national and local level[26] Since May the start of the OCV program to May 2018 over 25 million vaccines have been distributed to countries who meet the above criteria[26]
Treatment
The basic, overall treatment for Cholera is re-hydration, to replace the fluids that have been lost. Those with mild dehydration can be treated orally with an oral re hydration solution also known as, (ORS).[24] When patients are severely dehydrated and unable to take in the proper amount of ORS, IV fluid treatment is generally pursued. Antibiotics are used in some cases, typically fluoroquinolones and tetracyclines.[24]
Genome
V. cholerae has two circular chromosomes, together totalling 4 million base pairs of DNA sequence and 3,885 predicted genes.[27] The genes for cholera toxin are carried by CTXphi (CTXφ), a temperate bacteriophage inserted into the V. cholerae genome. CTXφ can transmit cholera toxin genes from one V. cholerae strain to another, one form of horizontal gene transfer. The genes for toxin coregulated pilus are coded by the Vibrio pathogenicity island (VPI). The entire genome of the virulent strain V. cholerae El Tor N16961 has been sequenced,[1] and contains two circular chromosomes.[6] Chromosome 1 has 2,961,149 base pairs with 2,770 open reading frames (ORF's) and chromosome 2 has 1,072,315 base pairs, 1,115 ORF's. The larger first chromosome contains the crucial genes for toxicity, regulation of toxicity, and important cellular functions, such as transcription and translation.[1]
The second chromosome is determined to be different from a plasmid or megaplasmid due to the inclusion of housekeeping and other essential genes in the genome, including essential genes for metabolism, heat-shock proteins, and 16S rRNA genes, which are ribosomal subunit genes used to track evolutionary relationships between bacteria. Also relevant in determining if the replicon is a chromosome is whether it represents a significant percentage of the genome, and chromosome 2 is 40% by size of the entire genome. And, unlike plasmids, chromosomes are not self-transmissible.[6] However, the second chromosome may have once been a megaplasmid because it contains some genes usually found on plasmids.[1]
V. cholerae contains a genomic island of pathogenicity and is lysogenized with phage DNA. That means that the genes of a virus were integrated into the bacterial genome and made the bacteria pathogenic. The molecular pathway involved in expression of virulence is discussed in the pathology and current research sections below.
Bacteriophage CTXφ
CTXφ (also called CTXphi) is a filamentous phage that contains the genes for cholera toxin. Infectious CTXφ particles are produced when V. cholerae infects humans. Phage particles are secreted from bacterial cells without lysis. When CTXφ infects V. cholerae cells, it integrates into specific sites on either chromosome. These sites often contain tandem arrays of integrated CTXφ prophage. In addition to the ctxA and ctxB genes encoding cholera toxin, CTXφ contains eight genes involved in phage reproduction, packaging, secretion, integration, and regulation. The CTXφ genome is 6.9 kb long.[28]
Ecology and epidemiology
The main reservoirs of V. cholerae are aquatic sources such as rivers, brackish waters, and estuaries, often in association with copepods or other zooplankton, shellfish, and aquatic plants.[29]
Cholera infections are most commonly acquired from drinking water in which V. cholerae is found naturally or into which it has been introduced from the feces of an infected person. Cholera is most likely to be found and spread in places with inadequate water treatment, poor sanitation, and inadequate hygiene. Other common vehicles include raw or undercooked fish and shellfish. Transmission from person to person is very unlikely, and casual contact with an infected person is not a risk for becoming ill.[30]V. cholerae thrives in an aquatic environment, particularly in surface water. The primary connection between humans and pathogenic strains is through water, particularly in economically reduced areas that do not have good water purification systems.[18]
Nonpathogenic strains are also present in water ecologies. The wide variety of pathogenic and nonpathogenic strains that co-exist in aquatic environments are thought to allow for so many genetic varieties. Gene transfer is fairly common amongst bacteria, and recombination of different V. cholerae genes can lead to new virulent strains.[31]
A symbiotic relationship between V. cholerae and Ruminococcus obeum has been determined. R. obeum autoinducer represses the expression of several V. cholerae virulence factors. This inhibitory mechanism is likely to be present in other gut microbiota species which opens the way to mine the gut microbiota of members in specific communities which may utilize autoinducers or other mechanisms in order to restrict colonization by V. cholerae or other enteropathogens.
Outbreaks of Cholera cause an estimated 120,000 deaths annually worldwide. There has been roughly seven pandemics since 1817, the first. These pandemics first arose in the Indian subcontinent and spread.[18]
Diversity and evolution
Two serogroups of V. cholerae, O1 and O139, cause outbreaks of cholera. O1 causes the majority of outbreaks, while O139 – first identified in Bangladesh in 1992 – is confined to Southeast Asia. Many other serogroups of V. cholerae, with or without the cholera toxin gene (including the nontoxigenic strains of the O1 and O139 serogroups), can cause a cholera-like illness. Only toxigenic strains of serogroups O1 and O139 have caused widespread epidemics.
V. cholerae O1 has two biotypes, classical and El Tor, and each biotype has two distinct serotypes, Inaba and Ogawa. The symptoms of infection are indistinguishable, although more people infected with the El Tor biotype remain asymptomatic or have only a mild illness. In recent years, infections with the classical biotype of V. cholerae O1 have become rare and are limited to parts of Bangladesh and India.[32] Recently, new variant strains have been detected in several parts of Asia and Africa. Observations suggest these strains cause more severe cholera with higher case fatality rates.
Natural genetic transformation
V. cholerae can be induced to become competent for natural genetic transformation when grown on chitin, a biopolymer that is abundant in aquatic habitats (e.g. from crustacean exoskeletons).[33] Natural genetic transformation is a sexual process involving DNA transfer from one bacterial cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. Transformation competence in V. cholerae is stimulated by increasing cell density accompanied by nutrient limitation, a decline in growth rate, or stress.[33] The V. cholerae uptake machinery involves a competence-induced pilus, and a conserved DNA binding protein that acts as a ratchet to reel DNA into the cytoplasm.[34] [35] There are two models of genetic transformation, sex hypothesis and competent bacteria.[36]
Gallery
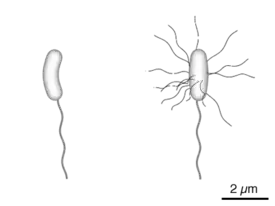 Diagram of the bacterium, V. cholerae
Diagram of the bacterium, V. cholerae Yellow colored (sucrose-fermenting) colonies of Vibrio cholerae on TCBS agar.
Yellow colored (sucrose-fermenting) colonies of Vibrio cholerae on TCBS agar.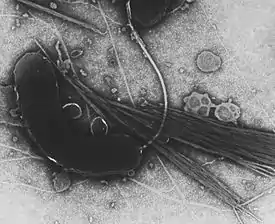 Transmission electron microscope image of Vibrio cholerae that has been negatively stained.
Transmission electron microscope image of Vibrio cholerae that has been negatively stained.
References
- "Laboratory Methods for the Diagnosis of Vibrio cholerae" (PDF). Centre for Disease Control. Retrieved 29 October 2013.
- See:
- Filippo Pacini (1854) "Osservazioni microscopiche e deduzioni patologiche sul cholera asiatico" (Microscopic observations and pathological deductions on Asiatic cholera), Gazzetta Medica Italiana: Toscana, 2nd series, 4 (50) : 397-401; 4 (51) : 405-412. The term "vibrio cholera" appears on page 411.
- Reprinted (more legibly) as a pamphlet.
- Real Academia de la Historia, ed. (2018). "Joaquín Balcells y Pasqual" (in Spanish). Archived from the original on 2019-07-08. Retrieved 2020-08-01.
- Col·legi Oficial de Metges de Barcelona, ed. (2015). "Joaquim Balcells i Pascual" (in Catalan). Archived from the original on 2020-08-01. Retrieved 2020-08-01.
- Bentivoglio, M; Pacini, P (1995). "Filippo Pacini: A determined observer" (PDF). Brain Research Bulletin. 38 (2): 161–5. CiteSeerX 10.1.1.362.6850. doi:10.1016/0361-9230(95)00083-Q. PMID 7583342. S2CID 6094598.
- Howard-Jones, N (1984). "Robert Koch and the cholera vibrio: a centenary". BMJ. 288 (6414): 379–81. doi:10.1136/bmj.288.6414.379. PMC 1444283. PMID 6419937.
- "New strains of Vibrio cholerae". www.mrc-lmb.cam.ac.uk. Retrieved 26 June 2019.
- Shigematsu, M.; Meno, Y.; Misumi, H.; Amako, K. (1995). "The measurement of swimming velocity of Vibrio cholerae and Pseudomonas aeruginosa using the video tracking methods". Microbiol Immunol. 39 (10): 741–4. doi:10.1111/j.1348-0421.1995.tb03260.x. PMID 8577263. S2CID 24759458.
- Jugder, Bat-Erdene; Watnick, Paula I. (2020-02-12). "Vibrio cholerae Sheds Its Coat to Make Itself Comfortable in the Gut". Cell Host & Microbe. 27 (2): 161–163. doi:10.1016/j.chom.2020.01.017. ISSN 1931-3128. PMID 32053783.
- Xi, Daoyi; Li, Yujia; Yan, Junxiang; Li, Yuehua; Wang, Xiaochen; Cao, Boyang (2020). "Small RNA coaR contributes to intestinal colonization in Vibrio cholerae via the two-component system EnvZ/OmpR". Environmental Microbiology. n/a (n/a): 4231–4243. doi:10.1111/1462-2920.14906. ISSN 1462-2920. PMID 31868254.
- Song, Tianyan; Mika, Franziska; Lindmark, Barbro; Liu, Zhi; Schild, Stefan; Bishop, Anne; Zhu, Jun; Camilli, Andrew; Johansson, Jörgen; Vogel, Jörg; Wai, Sun Nyunt (2008). "A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles". Molecular Microbiology. 70 (1): 100–111. doi:10.1111/j.1365-2958.2008.06392.x. ISSN 1365-2958. PMC 2628432. PMID 18681937.
- Bradley, Evan S.; Bodi, Kip; Ismail, Ayman M.; Camilli, Andrew (2011-07-14). "A Genome-Wide Approach to Discovery of Small RNAs Involved in Regulation of Virulence in Vibrio cholerae". PLOS Pathogens. 7 (7): e1002126. doi:10.1371/journal.ppat.1002126. ISSN 1553-7374. PMC 3136459. PMID 21779167.
- Davis, B; Waldor, M. K. (February 2003). "Filamentous phages linked to virulence of Vibrio cholerae". Current Opinion in Microbiology. 6 (1): 35–42. doi:10.1016/S1369-5274(02)00005-X. PMID 12615217.
- Boyd, EF; Waldor, MK (Jun 2002). "Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates". Microbiology. 148 (Pt 6): 1655–66. doi:10.1099/00221287-148-6-1655. PMID 12055286.
- Schillberg E., Ariti C., Bryson L., Delva-Senat R., Price D., GrandPierre , Lenglet A. (2016). "Factors Related to Fetal Death in Pregnant Women with Cholera, Haiti, 2011–2014". Emerging Infectious Diseases. 22 (1): 124–127. doi:10.3201/eid2201.151078. PMC 4696702. PMID 26692252.CS1 maint: multiple names: authors list (link)
- Miller, Melissa B.; Skorupski, Karen; Lenz, Derrick H.; Taylor, Ronald K.; Bassler, Bonnie L. (August 2002). "Parallel Quorum Sensing Systems Converge to Regulate Virulence in Vibrio cholerae". Cell. 110 (3): 303–314. doi:10.1016/S0092-8674(02)00829-2. PMID 12176318. S2CID 696469.
- Nielsen, Alex Toftgaard; Dolganov, Nadia A.; Otto, Glen; Miller, Michael C.; Wu, Cheng Yen; Schoolnik, Gary K. (2006). "RpoS Controls the Vibrio cholerae Mucosal Escape Response". PLOS Pathogens. 2 (10): e109. doi:10.1371/journal.ppat.0020109. PMC 1617127. PMID 17054394.
- Faruque, SM; Albert, MJ; Mekalanos, JJ (Dec 1998). "Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae". Microbiology and Molecular Biology Reviews. 62 (4): 1301–14. doi:10.1128/MMBR.62.4.1301-1314.1998. PMC 98947. PMID 9841673.
- "Illness & Symptoms | Cholera | CDC". www.cdc.gov. 2018-12-13. Retrieved 2019-11-12.
- “Cholera.” World Health Organization, World Health Organization, 17 Jan. 2019, www.who.int/news-room/fact-sheets/detail/cholera.
- “World Health Organization, Disease Outbreaks.” World Health Organization, World Health Organization, 8 Mar. 2016, www.searo.who.int/topics/disease_outbreaks/en/.
- “Mystery of Yemen Cholera Epidemic Solved.” ScienceDaily, ScienceDaily, 2 Jan. 2019, www.sciencedaily.com/releases/2019/01/190102140745.htm.
- "Five Basic Cholera Preventions". Center for Disease Control and Prevention. Retrieved 20 November 2019.
- Waldor and Edward T. Ryan. "Vibrio Matthew K. cholerae". doi:10.1016/B978-1-4557-4801-3.00216-2. Cite journal requires
|journal=(help)CS1 maint: uses authors parameter (link) - “Five Basic Cholera Prevention Steps | Cholera | CDC.” Centers for Disease Control and Prevention, Centers for Disease Control and Prevention, www.cdc.gov/cholera/preventionsteps.html.
- “Oral Cholera Vaccines.” World Health Organization, World Health Organization, 17 May 2018, www.who.int/cholera/vaccines/en/.
- Fraser, Claire M.; Heidelberg, John F.; Eisen, Jonathan A.; Nelson, William C.; Clayton, Rebecca A.; Gwinn, Michelle L.; Dodson, Robert J.; Haft, Daniel H.; et al. (2000). "DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae" (PDF). Nature. 406 (6795): 477–83. Bibcode:2000Natur.406..477H. doi:10.1038/35020000. PMID 10952301. S2CID 807509.
- McLeod, S. M.; Kimsey, H. H.; Davis, B. M.; Waldor, M. K. (2005). "CTXφ and Vibrio cholerae: exploring a newly recognized type of phage-host cell relationship". Molecular Microbiology. 57 (2): 347–356. doi:10.1111/j.1365-2958.2005.04676.x. PMID 15978069.
- Lutz, Carla; Erken, Martina; Noorian, Parisa; Sun, Shuyang; McDougald, Diane (2013). "Environmental reservoirs and mechanisms of persistence of Vibrio cholerae". Frontiers in Microbiology. 4: 375. doi:10.3389/fmicb.2013.00375. ISSN 1664-302X. PMC 3863721. PMID 24379807.
- "General Information | Cholera | CDC". www.cdc.gov. 2018-12-13. Retrieved 2019-11-14.
- Faruque, SM; Nair, GB (2002). "Molecular ecology of toxigenic Vibrio cholerae". Microbiology and Immunology. 46 (2): 59–66. doi:10.1111/j.1348-0421.2002.tb02659.x. PMID 11939579.
- Siddique, A.K.; Baqui, A.H.; Eusof, A.; Haider, K.; Hossain, M.A.; Bashir, I.; Zaman, K. (1991). "Survival of classic cholera in Bangladesh". The Lancet. 337 (8750): 1125–1127. doi:10.1016/0140-6736(91)92789-5. PMID 1674016. S2CID 33198972.
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK (2005). "Chitin induces natural competence in Vibrio cholerae". Science. 310 (5755): 1824–7. Bibcode:2005Sci...310.1824M. doi:10.1126/science.1120096. PMID 16357262. S2CID 31153549.
- Matthey N, Blokesch M (2016). "The DNA-Uptake Process of Naturally Competent Vibrio cholerae". Trends Microbiol. 24 (2): 98–110. doi:10.1016/j.tim.2015.10.008. PMID 26614677.
- quintdaily (3 August 2017). "Vibrio Cholera Starts Spreading In India – QuintDaily".
- Johnsborg, O; Eldholm, V; Ha˚varstein, L (2007). "Natural genetic transformation: prevalence, mechanisms and function". Department of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences, A˚ S, Norway. 158 (10): 767–778. doi:10.1016/j.resmic.2007.09.004. PMID 17997281.
External links
| Wikimedia Commons has media related to Vibrio cholerae. |
| Wikispecies has information related to Vibrio cholerae. |