Human mitochondrial DNA haplogroup
In human genetics, a human mitochondrial DNA haplogroup is a haplogroup defined by differences in human mitochondrial DNA. Haplogroups are used to represent the major branch points on the mitochondrial phylogenetic tree. Understanding the evolutionary path of the female lineage has helped population geneticists trace the matrilineal inheritance of modern humans back to human origins in Africa and the subsequent spread around the globe.
The letter names of the haplogroups (not just mitochondrial DNA haplogroups) run from A to Z. As haplogroups were named in the order of their discovery, the alphabetical ordering does not have any meaning in terms of actual genetic relationships.
The hypothetical woman at the root of all these groups (meaning just the mitochondrial DNA haplogroups) is the matrilineal most recent common ancestor (MRCA) for all currently living humans. She is commonly called Mitochondrial Eve.
The rate at which mitochondrial DNA mutates is known as the mitochondrial molecular clock. It is an area of ongoing research with one study reporting one mutation per 8000 years.[2]
Phylogeny
| Part of a series on |
| Genetic genealogy |
|---|
| Concepts |
| Related topics |
|
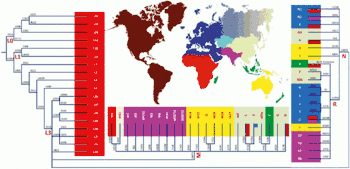
This phylogenetic tree is based Van Oven (2009).[4]
Chronology
| Haplogroup | Est. time of origin (kya)[5] | Possible place of origin | Highest frequencies |
|---|---|---|---|
| L | 200 | ||
| L1-6 | 170 | East Africa | |
| L2-6 | 150 | East Africa | |
| L0 | 150 | East Africa | |
| L1 | 140 | Central Africa | |
| L3-6 | 130 | ||
| L5 | 120 | ||
| L2 | 90 | ||
| L3 | 70 | East Africa | |
| N | 70 | East Africa or West Asia | |
| M | 60 | East Africa, West Asia or South Asia | |
| R | 60 | ||
| U | 55 | North-East Africa or India (South Asia) | |
| RT'JT | 55 | Middle East | |
| JT | 50 | Middle East | |
| U8 | 50 | Western Asia | |
| R9 | 47 | ||
| B4 | 44 | ||
| F | 43 | ||
| U4'9 | 42 | Central Asia | |
| U5 | 35 | Western Asia | |
| U6 | 35 | North Africa | |
| J | 35 | ||
| X | 30 | ||
| K | 30 | ||
| U5a | 27 | ||
| HV | 27 | Near East | |
| J1a | 27 | Near East | |
| T | 27 | Mesopotamia | |
| K1 | 27 | ||
| I | 26 | ||
| J1 | 24 | Near East | |
| W | 20 | ||
| U4 | 20 | Central Asia | |
| X2 | 20 | ||
| H | 20 | Western Asia | |
| U5a1 | 18 | Europe | |
| J1b | 11 | ||
| V | 14 | ||
| X2a | 13 | North America | |
| H1 | 12 | ||
| H3 | 12 | ||
| X1 | 10 | ||
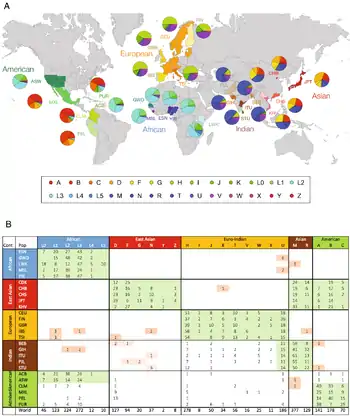
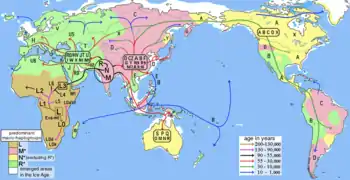
Geographical distribution
A 2004 paper suggested that the haplogroups most common in modern European populations were: H, J, K, N1, T, U4, U5, V, X and W.[6]
African haplogroups: L0, L1, L2, L3, L4, L5, L6, T, U5a
Australian haplogroups: M42a, M42c, M14, M15, Q, S, O, N, P. (Refs 1, 2, 3, 4, 5, 6)
Asian haplogroups: F, C, W, M, D, N, K, U, T, A, B, C, Z, U many number variants to each section
See also
References
- Rishishwar L, Jordan IK (2017). "Implications of human evolution and admixture for mitochondrial replacement therapy". BMC Genomics. 18 (1): 140. doi:10.1186/s12864-017-3539-3. PMC 5299762. PMID 28178941.
- Loogvali, Eva-Liis; Kivisild, Toomas; Margus, Tõnu; Villems, Richard (2009), O'Rourke, Dennis (ed.), "Explaining the Imperfection of the Molecular Clock of Hominid Mitochondria", PLOS ONE, 4 (12): e8260, Bibcode:2009PLoSO...4.8260L, doi:10.1371/journal.pone.0008260, PMC 2794369, PMID 20041137
- Kivisild T (2015). "Maternal ancestry and population history from whole mitochondrial genomes". Investig Genet. 6: 3. doi:10.1186/s13323-015-0022-2. PMC 4367903. PMID 25798216.
- van Oven M, Kayser M (February 2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–94. doi:10.1002/humu.20921. PMID 18853457. S2CID 27566749.
-
"Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock Supplementary" (PDF). 2009: 82–83 [89]. Archived from the original (PDF) on 2009-12-29. Cite journal requires
|journal=(help) - Villems, Richard; Usanga, Esien; Mikerezi, Ilia; Gölge, Mukaddes; Claustres, Mireille; Michalodimitrakis, Emmanuel N.; Pappa, Kalliopi I.; Anagnou, Nicholas P.; Chaventré, André; Moisan, Jean-Paul; Richard, Christelle; Grechanina, Elena; Balanovska, Elena V.; Rudan, Pavao; Puzyrev, Valery; Stepanov, Vadim; Khusnutdinova, Elsa K.; Gusar, Vladislava; Balanovsky, Oleg P.; Peričić, Marijana; Barać, Lovorka; Golubenko, Maria; Lunkina, Arina; Laos, Sirle; Pennarun, Erwan; Parik, Jüri; Tolk, Helle-Viivi; Reidla, Maere; Tambets, Kristiina; Metspalu, Ene; Kivisild, Toomas; Derenko, Miroslava V.; Malyarchuk, Boris A.; Roostalu, Urmas; Loogväli, Eva-Liis (November 1, 2004). "Disuniting Uniformity: A Pied Cladistic Canvas of mtDNA Haplogroup H in Eurasia". Molecular Biology and Evolution. 21 (11): 2012–2021. doi:10.1093/molbev/msh209. PMID 15254257 – via academic.oup.com.
External links
| Wikimedia Commons has media related to Human mtDNA haplogroups. |
- Mitochondrial phylogenetic trees
- Mannis van Oven's PhyloTree.org
- PhyloD3 – D3.js-based phylogenetic tree based on PhyloTree
- Mitochondrial haplogroup skeleton
- Vincent Macaulay's Mitochondrial haplogroup motifs
- List of mtDNA haplogroup projects
- MitoTool: a web server for the analysis and retrieval of human mitochondrial DNA sequence variations
- HaploGrep: mtDNA haplogroup determination based on PhyloTree.org
- HaploFind – fast automatic haplogroup assignment pipeline for human mitochondrial DNA
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||