RPGRIP1
X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 is a protein in the ciliary transition zone that in humans is encoded by the RPGRIP1 gene.[5][6] RPGRIP1 is a multi-domain protein containing a coiled-coil domain at the N-terminus, two C2 domains and a C-terminal RPGR-interacting domain (RID). Defects in the gene result in the Leber congenital amaurosis (LCA) syndrome[7] and in the eye disease glaucoma.[8]
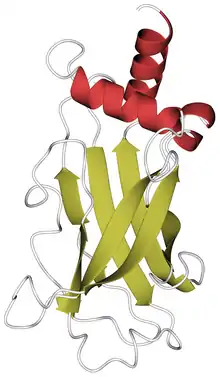 Crystal structure of the RPGR-interacting domain (RID) of RPGRIP1, PDB code 4qam. Alpha helices are in red, beta strands in gold. | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 | ||||||||
| Pfam | PF00168 | ||||||||
| InterPro | IPR031134 | ||||||||
| CATH | 4qam | ||||||||
| SCOP2 | 4qam / SCOPe / SUPFAM | ||||||||
| |||||||||
Interactions
RPGRIP1 has been shown to interact with Retinitis pigmentosa GTPase regulator.[9] RPGRIP1 interacts with RPGR via its RPGR-interacting domain (RID), which folds into a C2 domain architecture and interacts with RPGR at three different locations: A β strand of the RID interacting with the large loop of RPGR, at a hydrophobic interaction site, and via the N-terminal region of the RID.[10]
References
- GRCh38: Ensembl release 89: ENSG00000092200 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000057132 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Boylan JP, Wright AF (September 2000). "Identification of a novel protein interacting with RPGR". Human Molecular Genetics. 9 (14): 2085–93. doi:10.1093/hmg/9.14.2085. PMID 10958647.
- "Entrez Gene: RPGRIP1 retinitis pigmentosa GTPase regulator interacting protein 1".
- Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, Andréasson S, Berson EL (May 2001). "Null RPGRIP1 alleles in patients with Leber congenital amaurosis". American Journal of Human Genetics. 68 (5): 1295–8. doi:10.1086/320113. PMC 1226111. PMID 11283794.
- Fernández-Martínez L, Letteboer S, Mardin CY, Weisschuh N, Gramer E, Weber BH, Rautenstrauss B, Ferreira PA, Kruse FE, Reis A, Roepman R, Pasutto F (April 2011). "Evidence for RPGRIP1 gene as risk factor for primary open angle glaucoma". European Journal of Human Genetics. 19 (4): 445–51. doi:10.1038/ejhg.2010.217. PMC 3060327. PMID 21224891.
- Roepman R, Bernoud-Hubac N, Schick DE, Maugeri A, Berger W, Ropers HH, Cremers FP, Ferreira PA (September 2000). "The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors". Human Molecular Genetics. 9 (14): 2095–105. doi:10.1093/hmg/9.14.2095. PMID 10958648.
- Remans K, Bürger M, Vetter IR, Wittinghofer A (July 2014). "C2 domains as protein-protein interaction modules in the ciliary transition zone". Cell Reports. 8 (1): 1–9. doi:10.1016/j.celrep.2014.05.049. PMID 24981858.
Further reading
- Hong DH, Yue G, Adamian M, Li T (April 2001). "Retinitis pigmentosa GTPase regulator (RPGRr)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium". The Journal of Biological Chemistry. 276 (15): 12091–9. doi:10.1074/jbc.M009351200. PMID 11104772.
- Gerber S, Perrault I, Hanein S, Barbet F, Ducroq D, Ghazi I, Martin-Coignard D, Leowski C, Homfray T, Dufier JL, Munnich A, Kaplan J, Rozet JM (August 2001). "Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis". European Journal of Human Genetics. 9 (8): 561–71. doi:10.1038/sj.ejhg.5200689. PMID 11528500.
- Mavlyutov TA, Zhao H, Ferreira PA (August 2002). "Species-specific subcellular localization of RPGR and RPGRIP isoforms: implications for the phenotypic variability of congenital retinopathies among species". Human Molecular Genetics. 11 (16): 1899–907. doi:10.1093/hmg/11.16.1899. PMID 12140192.
- Hameed A, Abid A, Aziz A, Ismail M, Mehdi SQ, Khaliq S (August 2003). "Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy". Journal of Medical Genetics. 40 (8): 616–9. doi:10.1136/jmg.40.8.616. PMC 1735563. PMID 12920076.
- Shu X, Fry AM, Tulloch B, Manson FD, Crabb JW, Khanna H, Faragher AJ, Lennon A, He S, Trojan P, Giessl A, Wolfrum U, Vervoort R, Swaroop A, Wright AF (May 2005). "RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin". Human Molecular Genetics. 14 (9): 1183–97. doi:10.1093/hmg/ddi129. PMID 15772089.
- Lu X, Guruju M, Oswald J, Ferreira PA (May 2005). "Limited proteolysis differentially modulates the stability and subcellular localization of domains of RPGRIP1 that are distinctly affected by mutations in Leber's congenital amaurosis". Human Molecular Genetics. 14 (10): 1327–40. doi:10.1093/hmg/ddi143. PMC 1769350. PMID 15800011.
- Lu X, Ferreira PA (June 2005). "Identification of novel murine- and human-specific RPGRIP1 splice variants with distinct expression profiles and subcellular localization". Investigative Ophthalmology & Visual Science. 46 (6): 1882–90. doi:10.1167/iovs.04-1286. PMC 1769349. PMID 15914599.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (October 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Booij JC, Florijn RJ, ten Brink JB, Loves W, Meire F, van Schooneveld MJ, de Jong PT, Bergen AA (November 2005). "Identification of mutations in the AIPL1, CRB1, GUCY2D, RPE65, and RPGRIP1 genes in patients with juvenile retinitis pigmentosa". Journal of Medical Genetics. 42 (11): e67. doi:10.1136/jmg.2005.035121. PMC 1735944. PMID 16272259.
- Roepman R, Letteboer SJ, Arts HH, van Beersum SE, Lu X, Krieger E, Ferreira PA, Cremers FP (December 2005). "Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations". Proceedings of the National Academy of Sciences of the United States of America. 102 (51): 18520–5. doi:10.1073/pnas.0505774102. PMC 1317916. PMID 16339905.
- Jacobson SG, Cideciyan AV, Aleman TS, Sumaroka A, Schwartz SB, Roman AJ, Stone EM (May 2007). "Leber congenital amaurosis caused by an RPGRIP1 mutation shows treatment potential". Ophthalmology. 114 (5): 895–8. doi:10.1016/j.ophtha.2006.10.028. PMID 17306875.
- Arts HH, Doherty D, van Beersum SE, Parisi MA, Letteboer SJ, Gorden NT, Peters TA, Märker T, Voesenek K, Kartono A, Ozyurek H, Farin FM, Kroes HY, Wolfrum U, Brunner HG, Cremers FP, Glass IA, Knoers NV, Roepman R (July 2007). "Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome". Nature Genetics. 39 (7): 882–8. doi:10.1038/ng2069. PMID 17558407.
- Patil H, Guruju MR, Cho KI, Yi H, Orry A, Kim H, Ferreira PA (February 2012). "Structural and functional plasticity of subcellular tethering, targeting and processing of RPGRIP1 by RPGR isoforms". Biology Open. 1 (2): 140–60. doi:10.1242/bio.2011489. PMC 3507198. PMID 23213406.
External links
- PDBe-KB provides an overview of all the structure information available in the PDB for Human X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGRIP1)