Race and genetics
The relationship between race and genetics is relevant to the controversy concerning race classification. In everyday life, many societies classify populations into groups based on phenotypical traits and impressions of probable geographic ancestry and cultural identity—these are the groups usually called "races" in countries like the United States, Brazil, and South Africa. Patterns of variation of human genetic traits are generally clinal, with more abrupt shifts at places where steady gene flow is interrupted. The pattern of genetic variants tend to form larger regional clusters. Such a pattern can be explained by the expansion of the human population from Africa and serial genetic bottlenecks.[1] This causes genetic clusters to correlate statistically with population groups when a number of alleles are evaluated.[2][3][4]
| Race |
|---|
| Genetics and differences |
| Society |
| Race and... |
| Related topics |
|
Genetic analysis enables scientists to estimate the geographic ancestry of a person by using ancestry-informative markers, and by inference the probable racial category into which they will be classified in a given society. In that way there is a distinct statistical correlation between gene frequencies and racial categories. However, because all populations are genetically diverse, and because there is a complex relation between ancestry, genetic makeup and phenotype, and because racial categories are based on subjective evaluations of the traits, there is no specific gene that can be used to determine a person's race.[5][6][7]
Some genetic variants that contribute to the risk of complex diseases are differently distributed among human populations. It is debated whether self-identified race ought to be used by medical practitioners as a proxy for the probability that an individual possesses risk-related variants.[8][9] Such practice may result in false attribution of causality, stigmatisation of high-risk populations, or underestimation of risk for other populations.[10][11][12] There is also a large body of evidence that environmental risk factors for complex diseases track racial categories in the United States.[5]
Genetic variation
Genetic variation arises from mutations, from natural selection, migration between populations (gene flow) and from the reshuffling of genes through sexual reproduction.[13] Mutations lead to a change in the DNA structure, as the order of the bases are rearranged. Resultantly, different polypeptide proteins are coded. Some mutations may be positive and can help the individual survive more effectively in their environment. Variation is counteracted by natural selection and by genetic drift; note too the founder effect, when a small number of initial founders establish a population which hence starts with a correspondingly small degree of genetic variation. Epigenetic inheritance involves heritable changes in phenotype (appearance) or gene expression caused by mechanisms other than changes in the DNA sequence.
Human phenotypes are highly polygenic (dependent on interaction by many genes) and are influenced by environment as well as by genetics.
Nucleotide diversity is based on single mutations, single nucleotide polymorphisms (SNPs). The nucleotide diversity between humans is about 0.1 percent (one difference per one thousand nucleotides between two humans chosen at random). This amounts to approximately three million SNPs (since the human genome has about three billion nucleotides). There are an estimated ten million SNPs in the human population.
Research has shown that non-SNP (structural) variation accounts for more human genetic variation than single nucleotide diversity. Structural variation includes copy-number variation and results from deletions, inversions, insertions and duplications. It is estimated that approximately 0.4 percent of the genomes of unrelated people differ, apart from copy number. When copy-number variation is included, human-to-human genetic variation is estimated to be at least 0.5 percent.
Research methods
Trait, protein and gene studies
Early racial classification attempts measured surface traits, particularly skin color, hair color and texture, eye color, and head size and shape. (Measurements of the latter through craniometry were repeatedly discredited in the late 19th and mid-20th centuries.[14])
Biological adaptation plays a role in these bodily features and skin type. According to Luigi Luca Cavalli-Sforza, "From a scientific point of view, the concept of race has failed to obtain any consensus; none is likely, given the gradual variation in existence. It may be objected that the racial stereotypes have a consistency that allows even the layman to classify individuals. However, the major stereotypes, all based on skin color, hair color and form, and facial traits, reflect superficial differences that are not confirmed by deeper analysis with more reliable genetic traits and whose origin dates from recent evolution mostly under the effect of climate and perhaps sexual selection".[15][16][17][18][19][20]
A relative handful of genes accounts for the inherited factors shaping a person's appearance.[21][22] Humans have an estimated 19,000–20,000 human protein-coding genes.[23] Richard Sturm and David Duffy describe 11 genes that affect skin pigmentation and explain most variations in human skin color, the most significant of which are MC1R, ASIP, OCA2, and TYR.[24] There is evidence that as many as 16 different genes could be responsible for eye color in humans; however, the main two genes associated with eye color variation are OCA2 and HERC2, and both are localized in chromosome 15.[25]
Analysis of blood proteins
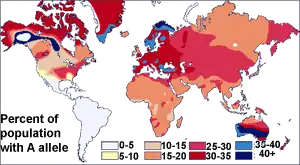
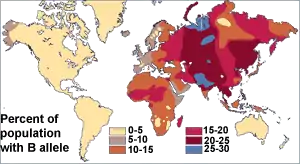
Before the discovery of DNA, scientists used blood proteins (the human blood group systems) to study human genetic variation. Research by Ludwik and Hanka Herschfeld during World War I found that the incidence of blood groups A and B differed by region; for example, among Europeans 15 percent were group B and 40 percent group A. Eastern Europeans and Russians had a higher incidence of group B; people from India had the greatest incidence. The Herschfelds concluded that humans comprised two "biochemical races", originating separately. It was hypothesized that these two races later mixed, resulting in the patterns of groups A and B. This was one of the first theories of racial differences to include the idea that human variation did not correlate with genetic variation. It was expected that groups with similar proportions of blood groups would be more closely related, but instead it was often found that groups separated by great distances (such as those from Madagascar and Russia), had similar incidences.[26] It was later discovered that the ABO blood group system is not just common to humans, but shared with other primates,[27] and likely predates all human groups.[28]
Population genetics
Researchers currently use genetic testing, which may involve hundreds (or thousands) of genetic markers or the entire genome.
Structure

Several methods to examine and quantify genetic subgroups exist, including cluster and principal components analysis. Genetic markers from individuals are examined to find a population's genetic structure. While subgroups overlap when examining variants of one marker only, when a number of markers are examined different subgroups have different average genetic structure. An individual may be described as belonging to several subgroups. These subgroups may be more or less distinct, depending on how much overlap there is with other subgroups.[29]
In cluster analysis, the number of clusters to search for K is determined in advance; how distinct the clusters are varies. The results obtained from cluster analyses depend on several factors:
- A large number genetic markers studied facilitates finding distinct clusters.[30]
- Some genetic markers vary more than others, so fewer are required to find distinct clusters.[2] Ancestry-informative markers exhibits substantially different frequencies between populations from different geographical regions. Using AIMs, scientists can determine a person's ancestral continent of origin based solely on their DNA. AIMs can also be used to determine someone's admixture proportions.[31]
- The more individuals studied, the easier it becomes to detect distinct clusters (statistical noise is reduced).[2]
- Low genetic variation makes it more difficult to find distinct clusters.[2] Greater geographic distance generally increases genetic variation, making identifying clusters easier.[32]
- A similar cluster structure is seen with different genetic markers when the number of genetic markers included is sufficiently large. The clustering structure obtained with different statistical techniques is similar. A similar cluster structure is found in the original sample with a subsample of the original sample.[3]
Recent studies have been published using an increasing number of genetic markers.[2][3][33][34][35][36]
Distance
Genetic distance is genetic divergence between species or populations of a species. It may compare the genetic similarity of related species, such as humans and chimpanzees. Within a species, genetic distance measures divergence between subgroups.
Genetic distance significantly correlates to geographic distance between populations, a phenomenon sometimes known as "isolation by distance".[37] Genetic distance may be the result of physical boundaries restricting gene flow such as islands, deserts, mountains or forests.
Genetic distance is measured by the fixation index (FST). FST is the correlation of randomly chosen alleles in a subgroup to a larger population. It is often expressed as a proportion of genetic diversity. This comparison of genetic variability within (and between) populations is used in population genetics. The values range from 0 to 1; zero indicates the two populations are freely interbreeding, and one would indicate that two populations are separate.
Many studies place the average FST distance between human races at about 0.125. Henry Harpending argued that this value implies on a world scale a "kinship between two individuals of the same human population is equivalent to kinship between grandparent and grandchild or between half siblings". In fact, the formulas derived in Harpending's paper in the "Kinship in a subdivided population" section imply that two unrelated individuals of the same race have a higher coefficient of kinship (0.125) than an individual and their mixed race half-sibling (0.109).[38]
History and geography
Cavalli-Sforza has described two methods of ancestry analysis.[39] Current-population genetic structure does not imply that differing clusters or components indicate only one ancestral home per group; for example, a genetic cluster in the US comprises Hispanics with European, Native American and African ancestry.[30]
Geographic analyses attempt to identify places of origin, their relative importance and possible causes of genetic variation in an area. The results can be presented as maps showing genetic variation. Cavalli-Sforza and colleagues argue that if genetic variations are investigated, they often correspond to population migrations due to new sources of food, improved transportation or shifts in political power. For example, in Europe the most significant direction of genetic variation corresponds to the spread of agriculture from the Middle East to Europe between 10,000 and 6,000 years ago.[39] Such geographic analysis works best in the absence of recent large-scale, rapid migrations.
Historic analyses use differences in genetic variation (measured by genetic distance) as a molecular clock indicating the evolutionary relation of species or groups, and can be used to create evolutionary trees reconstructing population separations.[39]
Validation
Results of genetic-ancestry research are supported if they agree with research results from other fields, such as linguistics or archeology.[39] Cavalli-Sforza and colleagues have argued that there is a correspondence between language families found in linguistic research and the population tree they found in their 1994 study. There are generally shorter genetic distances between populations using languages from the same language family. Exceptions to this rule are also found, for example Sami, who are genetically associated with populations speaking languages from other language families. The Sami speak a Uralic language, but are genetically primarily European. This is argued to have resulted from migration (and interbreeding) with Europeans while retaining their original language. Agreement also exists between research dates in archeology and those calculated using genetic distance.[2][39]
Group size
Research techniques can be used to detect genetic population differences if enough genetic markers are used; the Japanese and Chinese East Asian populations have been identified.[7] Sub-Saharan Africans have greater genetic diversity than other populations.[40]
Between-group genetics
In 1972, Richard Lewontin performed a FST statistical analysis using 17 markers (including blood-group proteins). He found that the majority of genetic differences between humans (85.4 percent) were found within a population, 8.3 percent were found between populations within a race and 6.3 percent were found to differentiate races (Caucasian, African, Mongoloid, South Asian Aborigines, Amerinds, Oceanians, and Australian Aborigines in his study). Since then, other analyses have found FST values of 6–10 percent between continental human groups, 5–15 percent between different populations on the same continent and 75–85 percent within populations.[41][42][43][44][45] This view has been affirmed by the American Anthropological Association and the American Association of Physical Anthropologists since.[46]
While acknowledging Lewontin's observation that humans are genetically homogeneous, A. W. F. Edwards in his 2003 paper "Human Genetic Diversity: Lewontin's Fallacy" argued that information distinguishing populations from each other is hidden in the correlation structure of allele frequencies, making it possible to classify individuals using mathematical techniques. Edwards argued that even if the probability of misclassifying an individual based on a single genetic marker is as high as 30 percent (as Lewontin reported in 1972), the misclassification probability nears zero if enough genetic markers are studied simultaneously. Edwards saw Lewontin's argument as based on a political stance, denying biological differences to argue for social equality.[47] Edwards' paper is reprinted, commented upon by experts such as Noah Rosenberg, and given further context in an interview with philosopher of science Rasmus Grønfeldt Winther in a recent anthology.[48]
As referred to before, Edwards criticises Lewontin's paper as he took 17 different traits and analysed them independently, without looking at them in conjunction with any other protein. Thus, it would have been fairly convenient for Lewontin to come up with the conclusion that racial naturalism is not tenable, according to his argument.[49] Sesardic also strengthened Edwards' view, as he used an illustration referring to squares and triangles, and showed that if you look at one trait in isolation, then it will most likely be a bad predicator of which group the individual belongs to.[50]
While acknowledging that FST remains useful, a number of scientists have written about other approaches to characterizing human genetic variation.[51][52][53] Long & Kittles (2009) stated that FST failed to identify important variation and that when the analysis includes only humans, FST = 0.119, but adding chimpanzees increases it only to FST = 0.183.[51] Mountain & Risch (2004) argued that an FST estimate of 0.10–0.15 does not rule out a genetic basis for phenotypic differences between groups and that a low FST estimate implies little about the degree to which genes contribute to between-group differences.[52] Pearse & Crandall 2004 wrote that FST figures cannot distinguish between a situation of high migration between populations with a long divergence time, and one of a relatively recent shared history but no ongoing gene flow.[53] In their 2015 article, Keith Hunley, Graciela Cabana, and Jeffrey Long (who had previously criticized Lewontin's statistical methodology with Rick Kittles[46]) recalculate the apportionment of human diversity using a more complex model than Lewontin and his successors. They conclude: "In sum, we concur with Lewontin's conclusion that Western-based racial classifications have no taxonomic significance, and we hope that this research, which takes into account our current understanding of the structure of human diversity, places his seminal finding on firmer evolutionary footing."[54]
Anthropologists (such as C. Loring Brace),[55] philosopher Jonathan Kaplan and geneticist Joseph Graves[56] have argued that while it is possible to find biological and genetic variation roughly corresponding to race, this is true for almost all geographically distinct populations: the cluster structure of genetic data is dependent on the initial hypotheses of the researcher and the populations sampled. When one samples continental groups, the clusters become continental; with other sampling patterns, the clusters would be different. Weiss and Fullerton note that if one sampled only Icelanders, Mayans and Maoris, three distinct clusters would form; all other populations would be composed of genetic admixtures of Maori, Icelandic and Mayan material.[57] Kaplan therefore concludes that, while differences in particular allele frequencies can be used to identify populations that loosely correspond to the racial categories common in Western social discourse, the differences are of no more biological significance than the differences found between any human populations (e.g., the Spanish and Portuguese).[58]
Self-identification
Jorde and Wooding found that while clusters from genetic markers were correlated with some traditional concepts of race, the correlations were imperfect and imprecise due to the continuous and overlapping nature of genetic variation, noting that ancestry, which can be accurately determined, is not equivalent to the concept of race.[7]
A 2005 study by Tang and colleagues used 326 genetic markers to determine genetic clusters. The 3,636 subjects, from the United States and Taiwan, self-identified as belonging to white, African American, East Asian or Hispanic ethnic groups. The study found "nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14 percent".[30]
Paschou et al. found "essentially perfect" agreement between 51 self-identified populations of origin and the population's genetic structure, using 650,000 genetic markers. Selecting for informative genetic markers allowed a reduction to less than 650, while retaining near-total accuracy.[59]
Correspondence between genetic clusters in a population (such as the current US population) and self-identified race or ethnic groups does not mean that such a cluster (or group) corresponds to only one ethnic group. African Americans have an estimated 20–25-percent European genetic admixture; Hispanics have European, Native American and African ancestry.[30] In Brazil there has been extensive admixture between Europeans, Amerindians and Africans. As a result, skin color differences within the population are not gradual, and there are relatively weak associations between self-reported race and African ancestry.[60][61] Ethnoracial self- classification in Brazilians is certainly not random with respect to genome individual ancestry, but the strength of the association between the phenotype and median proportion of African ancestry varies largely across population.[62]
Genetic-distance increase
Genetic distances generally increase continually with geographic distance, which makes a dividing line arbitrary. Any two neighboring settlements will exhibit some genetic difference from each other, which could be defined as a race. Therefore, attempts to classify races impose an artificial discontinuity on a naturally occurring phenomenon. This explains why studies on population genetic structure yield varying results, depending on methodology.[63]
Rosenberg and colleagues (2005) have argued, based on cluster analysis of the 52 populations in the Human Genetic Diversity Panel, that populations do not always vary continuously and a population's genetic structure is consistent if enough genetic markers (and subjects) are included.
Examination of the relationship between genetic and geographic distance supports a view in which the clusters arise not as an artifact of the sampling scheme, but from small discontinuous jumps in genetic distance for most population pairs on opposite sides of geographic barriers, in comparison with genetic distance for pairs on the same side. Thus, analysis of the 993-locus dataset corroborates our earlier results: if enough markers are used with a sufficiently large worldwide sample, individuals can be partitioned into genetic clusters that match major geographic subdivisions of the globe, with some individuals from intermediate geographic locations having mixed membership in the clusters that correspond to neighboring regions.
They also wrote, regarding a model with five clusters corresponding to Africa, Eurasia (Europe, Middle East, and Central/South Asia), East Asia, Oceania, and the Americas:
For population pairs from the same cluster, as geographic distance increases, genetic distance increases in a linear manner, consistent with a clinal population structure. However, for pairs from different clusters, genetic distance is generally larger than that between intracluster pairs that have the same geographic distance. For example, genetic distances for population pairs with one population in Eurasia and the other in East Asia are greater than those for pairs at equivalent geographic distance within Eurasia or within East Asia. Loosely speaking, it is these small discontinuous jumps in genetic distance—across oceans, the Himalayas, and the Sahara—that provide the basis for the ability of STRUCTURE to identify clusters that correspond to geographic regions.[3]
This applies to populations in their ancestral homes when migrations and gene flow were slow; large, rapid migrations exhibit different characteristics. Tang and colleagues (2004) wrote, "we detected only modest genetic differentiation between different current geographic locales within each race/ethnicity group. Thus, ancient geographic ancestry, which is highly correlated with self-identified race/ethnicity—as opposed to current residence—is the major determinant of genetic structure in the U.S. population".[30]
Number of clusters

Cluster analysis has been criticized because the number of clusters to search for is decided in advance, with different values possible (although with varying degrees of probability).[64] Principal component analysis does not decide in advance how many components for which to search.[65]
The 2002 study by Rosenberg et al.[66] exemplifies why meanings of these clusterings are disputable. The study shows that at the K=5 cluster analysis, genetic clusterings roughly map onto each of the five major geographical regions. Similar results were gathered in further studies in 2005.[67]
However, in addition to the five main supposedly geographical clusters, a sixth group, the Kalash, a minority ethnic group in Pakistan, began to appear starting at K=6. The racial naturalist Nicholas Wade considers that the results "make no genetic or geographic sense". They are therefore omitted in his book A Troublesome Inheritance in favour of the K=5 cluster analysis.
This bias, however, is reflective of how the research is inherently flawed. The sample population is chosen with geographical representation and folk concepts of race in mind, instead of accounting for the genetic diversity within the different geographical regions. The Kalash did not fit into the general pattern for it had been a genetically isolated population that happened to be reflected in this study. Potentially numerous genetically drifted groups, such as the uncontacted Sentinelese, are not represented in the study.
Utility
It has been argued that knowledge of a person's race is limited in value, since people of the same race vary from one another.[7] David J. Witherspoon and colleagues have argued that when individuals are assigned to population groups, two randomly chosen individuals from different populations can resemble each other more than a randomly chosen member of their own group. They found that many thousands of genetic markers had to be used for the answer to "How often is a pair of individuals from one population genetically more dissimilar than two individuals chosen from two different populations?" to be "never". This assumed three population groups, separated by large geographic distances (European, African and East Asian). The global human population is more complex, and studying a large number of groups would require an increased number of markers for the same answer. They conclude that "caution should be used when using geographic or genetic ancestry to make inferences about individual phenotypes",[68] and "The fact that, given enough genetic data, individuals can be correctly assigned to their populations of origin is compatible with the observation that most human genetic variation is found within populations, not between them. It is also compatible with our finding that, even when the most distinct populations are considered and hundreds of loci are used, individuals are frequently more similar to members of other populations than to members of their own population".[69]
This is similar to the conclusion reached by anthropologist Norman Sauer in a 1992 article on the ability of forensic anthropologists to assign "race" to a skeleton, based on craniofacial features and limb morphology. Sauer said, "the successful assignment of race to a skeletal specimen is not a vindication of the race concept, but rather a prediction that an individual, while alive was assigned to a particular socially constructed 'racial' category. A specimen may display features that point to African ancestry. In this country that person is likely to have been labeled Black regardless of whether or not such a race actually exists in nature".[70]
Ancestry-informative markers
Ancestry-informative markers (AIMs) are a genealogy tracing technology that has come under much criticism due to its reliance on reference populations. In a 2015 article, Troy Duster outlines how contemporary technology allows the tracing of ancestral lineage but along only the lines of one maternal and one paternal line. That is, of 64 total great-great-great-great-grandparents, only one from each parent is identified, implying the other 62 ancestors are ignored in tracing efforts.[71] Furthermore, the 'reference populations' used as markers for membership of a particular group are designated arbitrarily and contemporarily. In other words, using populations who currently reside in given places as references for certain races and ethnic groups is unreliable due to the demographic changes which have occurred over many centuries in those places. Furthermore, ancestry-informative markers being widely shared among the whole human population, it is their frequency which is tested, not their mere absence/presence. A threshold of relative frequency has, therefore, to be set. According to Duster the criteria for setting such thresholds are a trade secret of the companies marketing the tests. Thus, we cannot say anything conclusive on whether they are appropriate. Results of AIMs are extremely sensitive to where this bar is set.[72] Given that many genetic traits are found very similar amid many different populations, the rate of frequency which is taken to be enough for being part of a reference population is very important. This can also lead to mistakes, given that many populations may share the same patterns, if not exactly the same genes. "This means that someone from Bulgaria whose ancestors go back to the fifteenth century could (and sometime does) map as partly 'Native American'".[71] This happens because AIMs rely on a '100% purity' assumption of reference populations. That is, they assume that a pattern of traits would ideally be necessary and sufficient condition for assigning an individual to an ancestral reference populations.
Race and medicine
There are certain statistical differences between racial groups in susceptibility to certain diseases.[73] Genes change in response to local diseases; for example, people who are Duffy-negative tend to have a higher resistance to malaria. The Duffy negative phenotype is highly frequent in central Africa and the frequency decreases with distance away from Central Africa, with higher frequencies in global populations with high degrees of recent African immigration. This suggests that the Duffy negative genotype evolved in Sub-Saharan Africa and was subsequently positively selected for in the Malaria endemic zone.[74] A number of genetic conditions prevalent in malaria-endemic areas may provide genetic resistance to malaria, including sickle cell disease, thalassaemias and glucose-6-phosphate dehydrogenase. Cystic fibrosis is the most common life-limiting autosomal recessive disease among people of European ancestry; a hypothesized heterozygote advantage, providing resistance to diseases earlier common in Europe, has been challenged.[75] Scientists Michael Yudell, Dorothy Roberts, Rob DeSalle, and Sarah Tishkoff argue that using these associations in the practice of medicine has led doctors to overlook or misidentify disease: "For example, hemoglobinopathies can be misdiagnosed because of the identification of sickle-cell as a 'Black' disease and thalassemia as a 'Mediterranean' disease. Cystic fibrosis is underdiagnosed in populations of African ancestry, because it is thought of as a 'White' disease."[76]
Information about a person's population of origin may aid in diagnosis, and adverse drug responses may vary by group.[2] Because of the correlation between self-identified race and genetic clusters, medical treatments influenced by genetics have varying rates of success between self-defined racial groups.[77] For this reason, some physicians consider a patient's race in choosing the most effective treatment,[78] and some drugs are marketed with race-specific instructions.[79] Jorde and Wooding (2004) have argued that because of genetic variation within racial groups, when "it finally becomes feasible and available, individual genetic assessment of relevant genes will probably prove more useful than race in medical decision making". However, race continues to be a factor when examining groups (such as epidemiologic research).[7] Some doctors and scientists such as geneticist Neil Risch argue that using self-identified race as a proxy for ancestry is necessary to be able to get a sufficiently broad sample of different ancestral populations, and in turn to be able to provide health care that is tailored to the needs of minority groups.[80]
Usage in scientific journals
Some scientific journals have addressed previous methodological errors by requiring more rigorous scrutiny of population variables. Since 2000, Nature Genetics requires its authors to "explain why they make use of particular ethnic groups or populations, and how classification was achieved". Editors of Nature Genetics say that "[they] hope that this will raise awareness and inspire more rigorous designs of genetic and epidemiological studies".[81]
Gene-environment interactions
Lorusso and Bacchini[5] argue that self-identified race is of greater use in medicine as it correlates strongly with risk-related exposomes that are potentially heritable when they become embodied in the epigenome. They summarise evidence of the link between racial discrimination and health outcomes due to poorer food quality, access to healthcare, housing conditions, education, access to information, exposure to infectious agents and toxic substances, and material scarcity. They also cite evidence that this process can work positively – for example, the psychological advantage of perceiving oneself at the top of a social hierarchy is linked to improved health. However they caution that the effects of discrimination do not offer a complete explanation for differential rates of disease and risk factors between racial groups, and the employment of self-identified race has the potential to reinforce racial inequalities.
Objections to racial naturalism
Racial naturalism is the view that racial classifications are grounded in objective patterns of genetic similarities and differences. Proponents of this view have justified it using the scientific evidence described above. However, this view is controversial and philosopher[82]s of race have put forward four main objections to it.
Semantic objections, such as the discreteness objection, argue that the human populations picked out in population-genetic research are not races and do not correspond to what "race" means in the United States. "The discreteness objection does not require there to be no genetic admixture in the human species in order for there to be US 'racial groups' ... rather ... what the objection claims is that membership in US racial groups is different from membership in continental populations. ... Thus, strictly speaking, Blacks are not identical to Africans, Whites are not identical to Eurasians, Asians are not identical to East Asians and so forth."[83] Therefore, it could be argued that scientific research is not really about race.
The next two objections, are metaphysical objections which argue that even if the semantic objections fail, human genetic clustering results do not support the biological reality of race. The 'very important objection' stipulates that races in the US definition fail to be important to biology, in the sense that continental populations do not form biological subspecies. The 'objectively real objection' states that "US racial groups are not biologically real because they are not objectively real in the sense of existing independently of human interest, belief, or some other mental state of humans."[84] Racial naturalists, such as Quayshawn Spencer, have responded to each of these objections with counter-arguments. There are also methodological critics who reject racial naturalism because of concerns relating to the experimental design, execution, or interpretation of the relevant population-genetic research.[85]
Another semantic objection is the visibility objection which refutes the claim that there are US racial groups in human population structures. Philosophers such as Joshua Glasgow and Naomi Zack believe that US racial groups cannot be defined by visible traits, such as skin colour and physical attributes: "The ancestral genetic tracking material has no effect on phenotypes, or biological traits of organisms, which would include the traits deemed racial, because the ancestral tracking genetic material plays no role in the production of proteins it is not the kind of material that 'codes' for protein production."[86] Spencer contends that certain racial discourses require visible groups, but disagrees that this is a requirement in all US racial discourse.
A different objection states that US racial groups are not biologically real because they are not objectively real in the sense of existing independently of some mental state of humans. Proponents of this second metaphysical objection include Naomi Zack and Ron Sundstrom.[86][87] Spencer argues that an entity can be both biologically real and socially constructed. Spencer states that in order to accurately capture real biological entities, social factors must also be considered.
Criticism of race-based medicines
Troy Duster points out that genetics is often not the predominant determinant of disease susceptibilities, even though they might correlate with specific socially defined categories. This is because this research oftentimes lacks control for a multiplicity of socio-economic factors. He cites data collected by King and Rewers that indicates how dietary differences play a significant role in explaining variations of diabetes prevalence between populations.
Duster elaborates by putting forward the example of the Pima of Arizona, a population suffering from disproportionately high rates of diabetes. The reason for such, he argues, was not necessarily a result of the prevalence of the FABP2 gene, which is associated with insulin resistance. Rather he argues that scientists often discount the lifestyle implications under specific socio-historical contexts. For instance, near the end of the 19th century, the Pima economy was predominantly agriculture-based. However, as the European American population settles into traditionally Pima territory, the Pima lifestyles became heavily Westernised. Within three decades, the incidence of diabetes increased multiple folds. Governmental provision of free food to alleviate the prevalence of poverty in the population, which had comparatively high-fat contents, is noted as an explanation of this phenomenon.[88]
Lorusso and Bacchini argue against the assumption that "self-identified race is a good proxy for a specific genetic ancestry"[5] on the basis that self-identified race is complex: it depends on a range of psychological, cultural and social factors, and is therefore "not a robust proxy for genetic ancestry".[6] Furthermore, they explain that an individual's self-identified race is made up of further, collectively arbitrary factors: personal opinions about what race is and the extent to which it should be taken into consideration in everyday life. Furthermore, individuals who share a genetic ancestry may differ in their racial self-identification across historical or socioeconomic contexts. From this, Lorusso and Bacchini conclude that the accuracy in the prediction of genetic ancestry on the basis of self-identification is low, specifically in racially admixed populations born out of complex ancestral histories.
See also
- List of Y-chromosome haplogroups in populations of the world
- History of anthropometry, section; 4.2 Race, identity and cranio-facial description
- Human subspecies
References
- Kanitz, Ricardo; Guillot, Elsa G.; Antoniazza, Sylvain; Neuenschwander, Samuel; Goudet, Jérôme (2018-02-21). "Complex genetic patterns in human arise from a simple range-expansion model over continental landmasses". PLOS ONE. 13 (2): –0192460. Bibcode:2018PLoSO..1392460K. doi:10.1371/journal.pone.0192460. ISSN 1932-6203. PMC 5821356. PMID 29466398.
- Rosenberg NA; Pritchard JK; Weber JL; et al. (December 2002). "Genetic structure of human populations". Science. 298 (5602): 2381–5. Bibcode:2002Sci...298.2381R. doi:10.1126/science.1078311. PMID 12493913. S2CID 8127224.
- Rosenberg NA; Mahajan S; Ramachandran S; Zhao C; Pritchard JK; Feldman MW (December 2005). "Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure". PLOS Genetics. 1 (6): e70. doi:10.1371/journal.pgen.0010070. PMC 1310579. PMID 16355252.
- Tishkoff, Sarah A.; Reed, Floyd A.; Friedlaender, Françoise R.; Ehret, Christopher; Ranciaro, Alessia; Froment, Alain; Hirbo, Jibril B.; Awomoyi, Agnes A.; Bodo, Jean-Marie (2009-05-22). "The genetic structure and history of Africans and African Americans". Science. 324 (5930): 1035–1044. Bibcode:2009Sci...324.1035T. doi:10.1126/science.1172257. ISSN 1095-9203. PMC 2947357. PMID 19407144.
- Lorusso, Ludovica; Bacchini, Fabio (2015). "A reconsideration of the role of self-identified races in epidemiology and biomedical research". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 56–64. doi:10.1016/j.shpsc.2015.02.004. PMID 25791919.
- Hunt, L. M.; Megyesi, M.S. (Fall 2007). "The ambiguous meanings of the racial/ethnic categories routinely used in human genetics research". Social Science and Medicine. 66 (2): 349–361. doi:10.1016/j.socscimed.2007.08.034. PMC 2213883. PMID 17959289 – via Science Direct Assets.
- Jordge, Lynn B.; Wooding, Stephen P. (2004). "Genetic Variation, classification and 'race'". Nature Genetics. 36 (11 Suppl): S28–33. doi:10.1038/ng1435. PMID 15508000.
- Bustamante, Carlos D.; Burchard, Esteban González; De la Vega, Francisco M. (2011-07-13). "Genomics for the world". Nature. 475 (7355): 163–165. doi:10.1038/475163a. ISSN 1476-4687. PMC 3708540. PMID 21753830.
- Fejerman, Laura; Stern, Mariana C.; Ziv, Elad; John, Esther M.; Torres-Mejia, Gabriela; Hines, Lisa M.; Wolff, Roger; Wang, Wei; Baumgartner, Kathy B. (2013). "Genetic ancestry modifies the association between genetic risk variants and breast cancer risk among Hispanic and non-Hispanic white women". Carcinogenesis. 34 (8): 1787–1793. doi:10.1093/carcin/bgt110. ISSN 1460-2180. PMC 3731801. PMID 23563089.
- Roberts, Dorothy E. (2011-01-01). "Fatal Invention: How Science, Politics, and Big Business Re-create Race in the Twenty-first Century". Scholarship at Penn Law.
- Dorothy Roberts (14 June 2011). Fatal Invention: How Science, Politics, and Big Business Re-create Race in the Twenty-First Century. New Press/ORIM. ISBN 978-1-59558-691-9.
- Pepper, Michael; Stewart, Cheryl (2016). "Cystic fibrosis on the African continent". Genetics in Medicine. 18 (7): 653–662. doi:10.1038/gim.2015.157. hdl:2263/56176. ISSN 1530-0366. PMID 26656651.
- Livingstone, Frank (Summer 1962). "On the Non-Existence of Human Races" (PDF). Chicago Journals.
- Andrea Orsucci, ""Ariani, indogermani, stirpi mediterranee: aspetti del dibattito sulle razze europee (1870–1914)" Archived December 18, 2012, at Archive.today, Cromohs, 1998 (in Italian)
- Cavalli-Sforza, Luigi Luca; Menozzi, Paolo; Piazza, Alberto (1994). The History and Geography of Human Genes. Princeton: Princeton University Press. ISBN 978-0-691-08750-4. Lay summary (1 December 2013).
- McDonald, J; Lehman, DC (Spring 2012). "Forensic DNA analysis". Clinical Laboratory Science. 25 (2): 109–113. doi:10.29074/ascls.25.2.109. PMID 22693781.
- Butler, John. "Genetics and Genomics of Core STR Loci Used in Human Identity Testing*" (PDF).
- Redd, A. J.; Chamberlain, VF; Kearney, V. F.; Stover, D; Karafet, T.; Calderon, K.; Walsh, B.; Hammer, M. F. (May 2006). "Genetic structure among 38 populations from the United States based on 11 U.S. core Y chromosome STRs". Journal of Forensic Sciences. 51 (3): 580–585. doi:10.1111/j.1556-4029.2006.00113.x. PMID 16696705. S2CID 23597453.
- Hammer, M. F.; Chamberlain, V. F.; Kearney, V. F.; Stover, D.; Zhang, G; Karafet, T.; Walsh, B.; Redd, A. J. (December 1, 2006). "Population structure of Y chromosome SNP haplogroups in the United States and forensic implications for constructing Y chromosome STR databases". Forensic Science International. 164 (1): 45–55. doi:10.1016/j.forsciint.2005.11.013. PMID 16337103.
- Sims, L. M.; Ballantyne, J. (March 2008). "The golden gene (SLC24A5) differentiates US sub-populations within the ethnically admixed Y-SNP haplogroups" (PDF). Legal Medicine (Tokyo, Japan). 10 (2): 72–7. doi:10.1016/j.legalmed.2007.06.004. PMID 17720606.
- "Do Races Differ? Not Really, DNA Shows". The New York Times. 22 August 2000.
- Owens, Kelly; King, Mary-Claire (1999-10-15). "Genomic Views of Human History". Science. 286 (5439): 451–453. doi:10.1126/science.286.5439.451. ISSN 0036-8075. PMID 10521333.
Variation in other traits popularly used to identify 'races' is likely to be due to similarly straightforward mechanisms, involving limited numbers of genes with very specific physiological effects.
- Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML (November 2014). "Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes". Human Molecular Genetics. 23 (22): 5866–5878. doi:10.1093/hmg/ddu309. PMC 4204768. PMID 24939910.
- Sturm, Richard A.; Duffy, David L. (2012). "Human pigmentation genes under environmental selection". Genome Biology. 13 (9): 248. doi:10.1186/gb-2012-13-9-248. ISSN 1474-760X. PMC 3491390. PMID 23110848.
- White, Désirée; Rabago-Smith, Montserrat (January 2011). "Genotype-phenotype associations and human eye color". Journal of Human Genetics. 56 (1): 5–7. doi:10.1038/jhg.2010.126. PMID 20944644.
- Sykes, Bryan (2001). "From Blood Groups to Genes". The seven daughters of Eve. New York: Norton. pp. 32–51. ISBN 978-0-393-02018-2.
- Blancher, Antoine; Klein, Jan; Socha, Wladyslaw W. (2012). Molecular biology and evolution of blood group and MHC antigens in primates. Springer Science & Business Media. ISBN 978-3-642-59086-3.
- Ségurel, Laure; Thompson, Emma E.; Flutre, Timothée; Lovstad, Jessica; Venkat, Aarti; Margulis, Susan W.; Moyse, Jill; Ross, Steve; Gamble, Kathryn; Sella, Guy; Ober, Carole; Przeworski, Molly (2012-11-06). "The ABO blood group is a trans-species polymorphism in primates". Proceedings of the National Academy of Sciences. 109 (45): 18493–18498. arXiv:1208.4613. Bibcode:2012PNAS..10918493S. doi:10.1073/pnas.1210603109. ISSN 0027-8424. PMC 3494955. PMID 23091028.
- Witherspoon, D. J.; Wooding, S.; Rogers, A. R.; Marchani, E. E.; Watkins, W. S.; Batzer, M. A.; Jorde, L. B. (2007). "Genetic Similarities Within and Between Human Populations". Genetics. 176 (1): 351–359. doi:10.1534/genetics.106.067355. ISSN 0016-6731. PMC 1893020. PMID 17339205.
- Tang H, Quertermous T, Rodriguez B, et al. (February 2005). "Genetic Structure, Self-Identified Race/Ethnicity, and Confounding in Case-Control Association Studies". American Journal of Human Genetics. 76 (2): 268–75. doi:10.1086/427888. PMC 1196372. PMID 15625622.
- Lewontin, R. C. "Confusions About Human Races".
- Kittles RA, Weiss KM (2003). "Race, ancestry, and genes: implications for defining disease risk". Annual Review of Genomics and Human Genetics. 4: 33–67. doi:10.1146/annurev.genom.4.070802.110356. PMID 14527296.
- Li, J. Z.; Absher, D. M.; Tang, H.; Southwick, A. M.; Casto, A. M.; Ramachandran, S.; Cann, H. M.; Barsh, G. S.; Feldman, M.; Cavalli-Sforza, L. L.; Myers, R. M. (2008). "Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation". Science. 319 (5866): 1100–1104. Bibcode:2008Sci...319.1100L. doi:10.1126/science.1153717. PMID 18292342. S2CID 53541133.
- Jakobsson, M.; Scholz, S. W.; Scheet, P.; Gibbs, J. R.; Vanliere, J. M.; Fung, H. C.; Szpiech, Z. A.; Degnan, J. H.; Wang, K.; Guerreiro, R.; Bras, J. M.; Schymick, J. C.; Hernandez, D. G.; Traynor, B. J.; Simon-Sanchez, J.; Matarin, M.; Britton, A.; Van De Leemput, J.; Rafferty, I.; Bucan, M.; Cann, H. M.; Hardy, J. A.; Rosenberg, N. A.; Singleton, A. B. (2008). "Genotype, haplotype and copy-number variation in worldwide human populations" (PDF). Nature. 451 (7181): 998–1003. Bibcode:2008Natur.451..998J. doi:10.1038/nature06742. hdl:2027.42/62552. PMID 18288195. S2CID 11074384.
- Xing, J.; Watkins, W. S.; Witherspoon, D. J.; Zhang, Y.; Guthery, S. L.; Thara, R.; Mowry, B. J.; Bulayeva, K.; Weiss, R. B.; Jorde, L. B. (2009). "Fine-scaled human genetic structure revealed by SNP microarrays". Genome Research. 19 (5): 815–825. doi:10.1101/gr.085589.108. PMC 2675970. PMID 19411602.
- López Herráez, D.; Bauchet, M.; Tang, K.; Theunert, C.; Pugach, I.; Li, J.; Nandineni, M. R.; Gross, A.; Scholz, M.; Stoneking, M. (2009). Hawks, John (ed.). "Genetic Variation and Recent Positive Selection in Worldwide Human Populations: Evidence from Nearly 1 Million SNPs". PLOS ONE. 4 (11): e7888. Bibcode:2009PLoSO...4.7888L. doi:10.1371/journal.pone.0007888. PMC 2775638. PMID 19924308.
- Ramachandran, S; Deshpande, O; Roseman, CC; Rosenberg, NA; Feldman, MW; Cavalli-Sforza, LL (November 2005). "Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa". Proceedings of the National Academy of Sciences. 102 (44): 15942–7. Bibcode:2005PNAS..10215942R. doi:10.1073/pnas.0507611102. PMC 1276087. PMID 16243969.
- Harpending, Henry (2002-11-01). "Kinship and Population Subdivision" (PDF). Population & Environment. 24 (2): 141–147. doi:10.1023/A:1020815420693. JSTOR 27503827. S2CID 15208802.
- Luigi Luca Cavalli-Sforza, "Genes, peoples, and languages", Proceedings of the National Academy of Sciences, 1997, vol.94, pp.7719–7724, doi:10.1073/pnas.94.15.7719
- Long, Jeffrey C.; Li, Jie; Healy, Meghan E. (2009). "Human DNA sequences: More variation and less race" (PDF). American Journal of Physical Anthropology. 139 (1): 23–34. doi:10.1002/ajpa.21011. hdl:2027.42/62133. PMID 19226648.
- Lewontin, Richard (1972). "The Apportionment of Human Diversity". Evolutionary Biology. 6. pp. 381–398. doi:10.1007/978-1-4684-9063-3_14. ISBN 978-1-4684-9065-7. Missing or empty
|title=(help) - Risch, Neil; Burchard, Esteban; Ziv, Elad; Tang, Hua (2002). "Categorization of humans in biomedical research: genes, race and disease". Genome Biology. 3 (7): comment2007.1. doi:10.1186/gb-2002-3-7-comment2007. ISSN 1465-6906. PMC 139378. PMID 12184798.
- Templeton, Alan R. (2003). "Human Races in the Context of Recent Human Evolution: A Molecular Genetic Perspective". In Goodman, Alan H.; Heath, Deborah; Lindee, M. Susan (eds.). Genetic nature/culture: anthropology and science beyond the two-culture divide. Berkeley: University of California Press. pp. 234–257. ISBN 978-0-520-23792-6. Retrieved 23 September 2014.
- Ossorio P, Duster T (January 2005). "Race and genetics: controversies in biomedical, behavioral, and forensic sciences". The American Psychologist. 60 (1): 115–128. doi:10.1037/0003-066X.60.1.115. PMID 15641926.
- Lewontin, R. C. (2005). "Confusions About Human Races". Race and Genomics, Social Sciences Research Council. Retrieved 28 December 2006.
- Long, Jeffrey C.; Kittles, Rick A. (2009). "Human Genetic Diversity and the Nonexistence of Biological Races". Human Biology. 81 (5): 777–798. doi:10.3378/027.081.0621. ISSN 1534-6617. PMID 20504196. S2CID 30709062. Retrieved 2016-01-13.
- Edwards AW (August 2003). "Human genetic diversity: Lewontin's fallacy". BioEssays. 25 (8): 798–801. doi:10.1002/bies.10315. PMID 12879450.
- Winther, Rasmus Grønfeldt (2018). Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of A. W. F. Edwards with Commentaries. Cambridge, U.K.: Cambridge University Press. ISBN 9781107111721.
- Edwards, AWF (2003). Human genetic diversity: Lewontin's fallacy, BioEssays. pp. 798–801.
- Sesardic, N. (2010). Race: a social destruction of a biological concept. Biology and Philosophy. pp. 143–162.
- Long, J. C. & Kittles, R. A. (2009). "Human genetic diversity and the nonexistence of biological races". Human Biology. 81 (5/6): 777–798. doi:10.3378/027.081.0621. PMID 20504196. S2CID 30709062.
- Mountain, J. L. & Risch, N. (2004). "Assessing genetic contributions to phenotypic differences among 'racial' and 'ethnic' groups". Nature Genetics. 36 (11 Suppl): S48–S53. doi:10.1038/ng1456. PMID 15508003.
- Pearse, D. E.; Crandall, K. A. (2004). "Beyond FST: analysis of population genetic data for conservation". Conservation Genetics. 5 (5): 585–602. doi:10.1007/s10592-003-1863-4. S2CID 22068080.
- Hunley, Keith L.; Cabana, Graciela S.; Long, Jeffrey C. (2015-12-01). "The apportionment of human diversity revisited". American Journal of Physical Anthropology. 160 (4): 561–569. doi:10.1002/ajpa.22899. ISSN 1096-8644. PMID 26619959.
- Brace, C. Loring (2005). "Race" is a Four-letter Word: The Genesis of the Concept. Oxford: Oxford University Press. ISBN 978-0-19-517351-2. Lay summary (27 November 2010).
- Graves, Joseph L (2001). The Emperor's New Clothes: Biological Theories of Race at the Millennium. Rutgers University Press.
- Weiss, Kenneth M.; Fullerton, Stephanie M. (2005). "Racing around, getting nowhere". Evolutionary Anthropology: Issues, News, and Reviews. 14 (5): 165–169. doi:10.1002/evan.20079. ISSN 1060-1538. S2CID 84927946.
- Kaplan, Jonathan Michael (17 January 2011). "'Race': What Biology Can Tell Us about a Social Construct". Encyclopedia of Life Sciences (ELS). doi:10.1002/9780470015902.a0005857. ISBN 978-0470016176. Retrieved 23 September 2014.
- Paschou, Peristera; Lewis, Jamey; Javed, Asif; Drineas, Petros (2010). "Ancestry informative markers for fine-scale individual assignment to worldwide populations". J Med Genet. 47 (12): 835–847. doi:10.1136/jmg.2010.078212. PMID 20921023. S2CID 6432430.
- Pena, Sérgio D. J.; Di Pietro, Giuliano; Fuchshuber-Moraes, Mateus; Genro, Julia Pasqualini; Hutz, Mara H.; Kehdy, Fernanda de Souza Gomes; Kohlrausch, Fabiana; Magno, Luiz Alexandre Viana; Montenegro, Raquel Carvalho; Moraes, Manoel Odorico; de Moraes, Maria Elisabete Amaral; de Moraes, Milene Raiol; Ojopi, Élida B.; Perini, Jamila A.; Racciopi, Clarice; Ribeiro-dos-Santos, Ândrea Kely Campos; Rios-Santos, Fabrício; Romano-Silva, Marco A.; Sortica, Vinicius A.; Suarez-Kurtz, Guilherme (2011). Harpending, Henry (ed.). "The Genomic Ancestry of Individuals from Different Geographical Regions of Brazil is More Uniform Than Expected". PLoS ONE. 6 (2): e17063. Bibcode:2011PLoSO...617063P. doi:10.1371/journal.pone.0017063. PMC 3040205. PMID 21359226.
- Parra, F. C. (2002). "Color and genomic ancestry in Brazilians". Proceedings of the National Academy of Sciences. 100 (1): 177–182. Bibcode:2002PNAS..100..177P. doi:10.1073/pnas.0126614100. PMC 140919. PMID 12509516.
- Lima-Costa, M. Fernanda; Rodrigues, Laura C.; Barreto, Maurício L.; Gouveia, Mateus; Horta, Bernardo L.; Mambrini, Juliana; Kehdy, Fernanda S. G.; Pereira, Alexandre; Rodrigues-Soares, Fernanda; Victora, Cesar G.; Tarazona-Santos, Eduardo; Cesar, Cibele C.; Conceição, Jackson S.; Costa, Gustavo N. O.; Esteban, Nubia; Fiaccone, Rosemeire L.; Figueiredo, Camila A.; Firmo, Josélia O. A.; Horimoto, Andrea R. V. R.; Leal, Thiago P.; Machado, Moara; Magalhães, Wagner C. S.; De Oliveira, Isabel Oliveira; Peixoto, Sérgio V.; Rodrigues, Maíra R.; Santos, Hadassa C.; Silva, Thiago M. (2015). "Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative)". Scientific Reports. 5: 9812. Bibcode:2015NatSR...5E9812.. doi:10.1038/srep09812. PMC 5386196. PMID 25913126.
- Reanne Frank, "Back with a Vengeance: the Reemergence of a Biological Conceptualization of Race in Research on Race/Ethnic Disparities in Health" Archived 2008-12-01 at the Wayback Machine
- Bolnick, Deborah A. (2008). "Individual Ancestry Inference and the Reification of Race as a Biological Phenomenon". In Koenig, Barbara A.; Richardson, Sarah S.; Lee, Sandra Soo-Jin (eds.). Revisiting race in a genomic age. Rutgers University Press. ISBN 978-0-8135-4324-6.
- Patterson, Nick; Price, Alkes L.; Reich, David (2006). "Population Structure and Eigenanalysis". PLOS Genetics. 2 (12): e190. doi:10.1371/journal.pgen.0020190. PMC 1713260. PMID 17194218.
- Rosenberg; et al. (2002). Genetic Structure of Human Populations (Report).
- Rosenberg, N. A.; Mahajan, S.; Ramachandran, S.; Zhao, C.; Pritchard, J. K.; et al. (2005). "Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure". PLOS Genet. 1 (6): e70. doi:10.1371/journal.pgen.0010070. PMC 1310579. PMID 16355252.
- Witherspoon DJ, Wooding S, Rogers AR, et al. (May 2007). "Genetic Similarities Within and Between Human Populations". Genetics. 176 (1): 351–9. doi:10.1534/genetics.106.067355. PMC 1893020. PMID 17339205.
- Witherspoon DJ, Wooding S, Rogers AR, et al. (May 2007). "Genetic Similarities Within and Between Human Populations". Genetics. 176 (1): 358. doi:10.1534/genetics.106.067355. PMC 1893020. PMID 17339205.
- Sauer, N. J. (January 1992). "Forensic anthropology and the concept of race: if races don't exist, why are forensic anthropologists so good at identifying them?". Social Science & Medicine. 34 (2): 107–111. doi:10.1016/0277-9536(92)90086-6. PMID 1738862.
- Duster, Troy (March 2015). "A post-genomic surprise. The molecular reinscription of race in science, law and medicine". The British Journal of Sociology. 66 (1): 1–27. doi:10.1111/1468-4446.12118. ISSN 0007-1315. PMID 25789799.
- Fullwiley, D. (2008). "The Biologistical Construction of Race: 'Admixture' Technology and the New Genetic Medicine". Social Studies of Science, 38(5), 695–735. doi:10.1177/0306312708090796
- Risch N (July 2005). "The whole side of it--an interview with Neil Risch by Jane Gitschier". PLOS Genetics. 1 (1): e14. doi:10.1371/journal.pgen.0010014. PMC 1183530. PMID 17411332.
- Malaria and the Red Cell Archived 2011-11-27 at the Wayback Machine, Harvard University. 2002.
- Högenauer C, Santa Ana CA, Porter JL, et al. (December 2000). "Active intestinal chloride secretion in human carriers of cystic fibrosis mutations: an evaluation of the hypothesis that heterozygotes have subnormal active intestinal chloride secretion". Am. J. Hum. Genet. 67 (6): 1422–1427. doi:10.1086/316911. PMC 1287919. PMID 11055897.
- Yudell, Michael; Roberts, Dorothy; DeSalle, Rob; Tishkoff, Sarah (2016). "Taking Race out of Human Genetics". Science. 351 (6273): 564–65. doi:10.1126/science.aac4951. PMID 26912690. S2CID 206639306.
- Schwartz, Robert S. (2001). "Racial Differences in the Response to Drugs — Pointers to Genetic Differences". New England Journal of Medicine. 344 (18): 1393–1396. doi:10.1056/NEJM200105033441810. PMID 11333999.
- Bloche, Gregg M. (2004). "Race-Based Therapeutics". New England Journal of Medicine. 351 (20): 2035–2037. doi:10.1056/nejmp048271. PMID 15533852.
- Drug information for the drug Crestor. Warnings for this drug state, "People of Asian descent may absorb rosuvastatin at a higher rate than other people. Make sure your doctor knows if you are Asian. You may need a lower than normal starting dose."
- Risch, N.; Burchard, E.; Ziv, E.; Tang, H. (2002). "Categorization of humans in biomedical research: genes, race and disease". Genome Biol. 3 (7): 1–12. doi:10.1186/gb-2002-3-7-comment2007. PMC 139378. PMID 12184798.
- "Census, Race and Science". Nature Genetics. 24 (2): 97–98. 2000. doi:10.1038/72884. PMID 10655044.
- "Revelations", Shut Up and Listen, Palgrave Macmillan, ISBN 978-0-230-36298-7, retrieved 2021-01-28
- Spencer, Quayshawn (2015). "Philosophy of race meets population genetics". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 49. doi:10.1016/j.shpsc.2015.04.003. PMID 25963045.
- Spencer, Quayshawn (2015). "Philosophy of race meets population genetics". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 51. doi:10.1016/j.shpsc.2015.04.003. PMID 25963045.
- Spencer, Quayshawn (2015). "Philosophy of race meets population genetics". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 46–47. doi:10.1016/j.shpsc.2015.04.003. PMID 25963045.
- Zach, Naomi (2002). "Philosophy of Science and Race". The International Journal of African Historical Studies. 36 (3): 656–658. doi:10.2307/3559445. JSTOR 3559445.
- Sundstrom, R (2002). "Race as a human kind". Philosophy & Social Criticism. 28: 91–115. doi:10.1177/0191453702028001592. S2CID 145381236.
- Duster, Troy (2015). "A post-genomic surprise. The molecular reinscription of race in science, law and medicine". The British Journal of Sociology. 66 (1): 1–27. doi:10.1111/1468-4446.12118. PMID 25789799.
Further reading
- Helms JE, Jernigan M, Mascher J (January 2005). "The meaning of race in psychology and how to change it: a methodological perspective". The American Psychologist. 60 (1): 27–36. doi:10.1037/0003-066X.60.1.27. PMID 15641919.
- Keita SO, Kittles RA, Royal CD, et al. (November 2004). "Conceptualizing human variation". Nature Genetics. 36 (11 Suppl): S17–20. doi:10.1038/ng1455. PMID 15507998.
- Koenig, Barbara A.; Lee, Sandra Soo-jin; Richardson, Sarah S., eds. (2008). Revisiting Race in a Genomic Age. New Brunswick (NJ): Rutgers University Press. ISBN 978-0-8135-4324-6. Lay summary (24 November 2010). This review of current research includes chapters by Jonathan Marks, John Dupré, Sally Haslanger, Deborah A. Bolnick, Marcus W. Feldman, Richard C. Lewontin, Sarah K. Tate, David B. Goldstein, Jonathan Kahn, Duana Fullwiley, Molly J. Dingel, Barbara A. Koenig, Mark D. Shriver, Rick A. Kittles, Henry T. Greely, Kimberly Tallbear, Alondra Nelson, Pamela Sankar, Sally Lehrman, Jenny Reardon, Jacqueline Stevens, and Sandra Soo-Jin Lee.
- Lieberman, Leonard; Kirk, Rodney C.; Corcoran, Michael (2003). "The Decline of Race in American Physical Anthropology" (PDF). Przegląd Antropologiczny – Anthropological Review. 66: 3–21. ISSN 0033-2003. Archived from the original (PDF) on 2011-06-08. Retrieved 2010-09-12.
- Long JC, Kittles RA (August 2003). "Human genetic diversity and the nonexistence of biological races". Human Biology. 75 (4): 449–71. doi:10.1353/hub.2003.0058. PMID 14655871. S2CID 26108602.
- Miththapala, Sriyanie; Seidensticker, John; O'Brien, Stephen J. (1996). "Phylogeographic Subspecies Recognition in Leopards (Panthera pardus): Molecular Genetic Variation". Conservation Biology. 10 (4): 1115–1132. doi:10.1046/j.1523-1739.1996.10041115.x.
- Ossorio P, Duster T (January 2005). "Race and genetics: controversies in biomedical, behavioral, and forensic sciences". The American Psychologist. 60 (1): 115–28. doi:10.1037/0003-066X.60.1.115. PMID 15641926.
- Parra EJ, Kittles RA, Shriver MD (November 2004). "Implications of correlations between skin color and genetic ancestry for biomedical research". Nature Genetics. 36 (11 Suppl): S54–60. doi:10.1038/ng1440. PMID 15508005.
- Sawyer SL, Mukherjee N, Pakstis AJ, et al. (May 2005). "Linkage disequilibrium patterns vary substantially among populations". European Journal of Human Genetics. 13 (5): 677–86. doi:10.1038/sj.ejhg.5201368. PMID 15657612.
- Rohde DL, Olson S, Chang JT (September 2004). "Modelling the recent common ancestry of all living humans". Nature. 431 (7008): 562–6. Bibcode:2004Natur.431..562R. CiteSeerX 10.1.1.78.8467. doi:10.1038/nature02842. PMID 15457259. S2CID 3563900.
- Serre D, Pääbo S (September 2004). "Evidence for Gradients of Human Genetic Diversity Within and Among Continents". Genome Research. 14 (9): 1679–85. doi:10.1101/gr.2529604. PMC 515312. PMID 15342553.
- Smedley A, Smedley BD (January 2005). "Race as biology is fiction, racism as a social problem is real: Anthropological and historical perspectives on the social construction of race". The American Psychologist. 60 (1): 16–26. CiteSeerX 10.1.1.568.4548. doi:10.1037/0003-066X.60.1.16. PMID 15641918.

