Species complex
In biology, a species complex is a group of closely related organisms that are so similar in appearance that the boundaries between them are often unclear. Terms that are sometimes used synonymously but have more precise meanings are cryptic species for two or more species hidden under one species name, sibling species for two cryptic species that are each other's closest relative, and species flock for a group of closely related species that live in the same habitat. As informal taxonomic ranks, species group, species aggregate, macrospecies, and superspecies are also in use.
Two or more taxa that were once considered conspecific (of the same species) may later be subdivided into infraspecific taxa (taxa within a species, such as bacterial strains or plant varieties), that is complex but it is not a species complex.
A species complex is in most cases a monophyletic group with a common ancestor, but there are exceptions. It may represent an early stage after speciation but may also have been separated for a long time period without evolving morphological differences. Hybrid speciation can be a component in the evolution of a species complex.
Species complexes exist in all groups of organisms and are identified by the rigorous study of differences between individual species that uses minute morphological details, tests of reproductive isolation, or DNA-based methods, such as molecular phylogenetics and DNA barcoding. The existence of extremely similar species may cause local and global species diversity to be underestimated. The recognition of similar-but-distinct species is important for disease and pest control and in conservation biology although the drawing of dividing lines between species can be inherently difficult.
Definition
A species complex is typically considered as a group of close, but distinct species.[5] Obviously, the concept is closely tied to the definition of a species. Modern biology understands a species as "separately evolving metapopulation lineage" but acknowledges that the criteria to delimit species may depend on the group studied.[6] Thus, many traditionally defined species defined, based only on morphological similarity, have been found to be several distinct species when other criteria, such as genetic differentiation or reproductive isolation, are applied.[7]
A more restricted use applies the term to close species between which hybridisation occurred or is occurring, which leads to intermediate forms and blurred species boundaries.[8] The informal classification, superspecies, can be exemplified by the grizzled skipper butterfly, which is a superspecies that is further divided into three subspecies.[9]
Some authors apply the term to a species with intraspecific variability, which might be a sign of ongoing or incipient speciation. Examples are ring species[10][11] or species with subspecies, in which it is often unclear if they should be considered separate species.[12]
Related concepts
Several terms are used synonymously for a species complex, but some of them may also have slightly different or narrower meanings. In the nomenclature codes of zoology and bacteriology, no taxonomic ranks are defined at the level between subgenera and species,[13][14] but the botanical code defines four ranks below genera (section, subsections, series and subseries).[15] Different informal taxonomic solutions have been used to indicate a species complex.
- Cryptic species
- Also called physiologic race[16] (uncommon). This describes "distinct species that are erroneously classified (and hidden) under one species name".[17] More generally, the term is often applied when species, even if they are known to be distinct, cannot be reliably distinguished by morphology. The usage physiologic race is not to be confused with physiological race.
- Sibling species
- Also called aphanic species. This term, introduced by Ernst Mayr in 1942,[18] was initially used with the same meaning as cryptic species,[7] but later authors emphasized the common phylogenetic origin.[19] A recent article defines sibling species as "cryptic sister species", "two species that are the closest relative of each other and have not been distinguished from one another taxonomically".[17]
- Species flock
- Also called species swarm. This refers to "a monophyletic group of closely related species all living in the same ecosystem".[17] Conversely, the term has also been applied very broadly to a group of closely related species than can be variable and widespread.[20] Not to be confused with a Mixed-species foraging flock, a behavior in which birds of different species feed together.
- Superspecies
- Sometimes used as an informal rank for a species complex around one "representative" species.[21][22] Popularized by Bernhard Rensch and later Ernst Mayr, with the initial requirement that species forming a superspecies must have allopatric distributions.[23] For the component species of a superspecies, allospecies was proposed.[23]
- Species aggregate
- Used for a species complex, especially in plant taxa where polypoidy and apomixis are common. Historical synonyms are species collectiva, introduced by Adolf Engler, conspecies, and grex.[24] Components of a species aggregate have been called segregates or microspecies.[24] Used as abbreviation agg. after the binomial species name.[8][25]
- Sensu lato
- A Latin phrase meaning "in the broad sense", it is often used after a binomial species name, often abbreviated as s.l., to indicate a species complex represented by that species.[26][27][28]
Identification
Distinguishing close species within a complex requires the study of often very small differences. Morphological differences may be minute and visible only by the use of adapted methods, such as microscopy. However, distinct species sometimes have no morphological differences.[17] In those cases, other characters, such as in the species' life history, behavior, physiology, and karyology, may be explored. For example, territorial songs are indicative of species in the treecreepers, a bird genus with few morphological differences.[29] Mating tests are common in some groups such as fungi to confirm the reproductive isolation of two species.[27]
Analysis of DNA sequences is becoming increasingly standard for species recognition and may, in many cases, be the only useful method.[17] Different methods are used to analyse such genetic data, such as molecular phylogenetics or DNA barcoding. Such methods have greatly contributed to the discovery of cryptic species,[17][30] including such emblematic species as the fly agaric[2] or the African elephants.[3]
Evolution and ecology
Speciation process
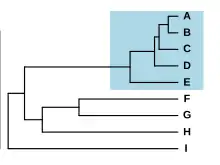
Species forming a complex have typically diverged very recently from each other, which sometimes allows the retracing of the process of speciation. Species with differentiated populations, such as ring species, are sometimes seen as an example of early, ongoing speciation: a species complex in formation. Nevertheless, similar but distinct species have sometimes been isolated for a long time without evolving differences, a phenomenon known as "morphological stasis".[17] For example, the Amazonian frog Pristimantis ockendeni is actually at least three different species that diverged over 5 million years ago.[32]
Stabilizing selection has been invoked as a force maintaining similarity in species complexes, especially they adaptat to special environments, such as a host in the case of symbionts or extreme environments, constrains possible directions of evolution: in such cases, strongly divergent selection is not to be expected.[17] Also, asexual reproduction, such as through apomixis in plants, may separate lineages without producing a great degree of morphological differentiation.
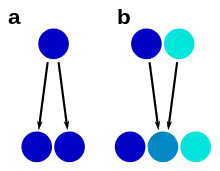
a – morphological stasis
b – hybrid speciation
A species complex is usually a group that has one common ancestor (a monophyletic group), but closer examination can sometimes disprove that. For example, yellow-spotted "fire salamanders" in the genus Salamandra, formerly all classified as one species S. salamandra, are not monophyletic: the Corsican fire salamander's closest relative has been shown to be the entirely black Alpine salamander.[31] In such cases, similarity has arisen from convergent evolution.
Hybrid speciation can lead to unclear species boundaries through a process of reticulate evolution, in which species have two parent species as their most recent common ancestors. In such cases, the hybrid species may have intermediate characters, such as in Heliconius butterflies.[33] Hybrid speciation has been observed in various species complexes, such as insects, fungi and plants. In plants, hybridization often takes place through polyploidization, and hybrid plant species are called nothospecies.
Range and habitats
Sources differ on whether or not members of a species group share a range. A source from Iowa State University Department of Agronomy states that members of a species group usually have partially overlapping ranges but do not interbreed with one another.[34] A Dictionary of Zoology (Oxford University Press 1999) describes a species group as complex of related species that exist allopatrically and explains that the "grouping can often be supported by experimental crosses in which only certain pairs of species will produce hybrids."[35] The examples given below may support both uses of the term "species group."
Often, such complexes do not become evident until a new species is introduced into the system, which breaks down existing species barriers. An example is the introduction of the Spanish slug in Northern Europe, where interbreeding with the local black slug and red slug, which were traditionally considered clearly separate species that did not interbreed, shows that they may be actually just subspecies of the same species.[36]
Where closely related species co-exist in sympatry, it is often a particular challenge to understand how the similar species persist without outcompeting each other. Niche partitioning is one mechanism invoked to explain that. Indeed, studies in some species complexes suggest that species divergence have gone in par with ecological differentiation, with species now preferring different microhabitats. Similar methods also found that the Amazonian frog Eleutherodactylus ockendeni is actually at least three different species that diverged over 5 million years ago.[32]
A species flock may arise when a species penetrates a new geographical area and diversifies to occupy a variety of ecological niches, a process known as adaptive radiation. The first species flock to be recognized as such was the 13 species of Darwin's finches on the Galápagos Islands described by Charles Darwin.
Practical implications
Biodiversity estimates
It has been suggested that cryptic species complexes are very common in the marine environment.[37] That suggestion came before the detailed analysis of many systems using DNA sequence data but has been proven to be correct.[38] The increased use of DNA sequence in the investigation of organismal diversity (also called phylogeography and DNA barcoding) has led to the discovery of a great many cryptic species complexes in all habitats. In the marine bryozoan Celleporella hyalina,[39] detailed morphological analyses and mating compatibility tests between the isolates identified by DNA sequence analysis were used to confirm that these groups consisted of more than 10 ecologically distinct species, which had been diverging for many million years.
Evidence from the identification of cryptic species has led some to conclude that current estimates of global species richness are too low.
Disease and pathogen control

Pests, species that cause diseases and their vectors, have direct importance for humans. When they are found to be cryptic species complexes, the ecology and the virulence of each of these species need to be re-evaluated to devise appropriate control strategies. Examples are cryptic species in the malaria vector genus of mosquito, Anopheles,the fungi causing cryptococcosis, and sister species of Bactrocera tyroni, or the Queensland fruit fly. Curiously, that pest is indistinguishable from two sister species except that B. tyroni inflicts widespread, devastating damage to Australian fruit crops, but the sister species do not.[41]
Conservation biology
When a species is found to be several phylogenetically distinct species, each typically has smaller distribution ranges and population sizes than had been reckoned. The different species can also differ in their ecology, such as by having different breeding strategies or habitat requirements, which must taken into account for appropriate management. For example, giraffe populations and subspecies differ genetically to such an extent that they may be considered species. Although the giraffe, as a whole, is not considered to be threatened, if each cryptic species is considered separately, there is a much higher level of threat.[42]
See also
References
- Ron, Santiago; Caminer, Marcel (2014). "Systematics of treefrogs of the Hypsiboas calcaratus and Hypsiboas fasciatus species complex (Anura, Hylidae) with the description of four new species". ZooKeys (370): 1–68. doi:10.3897/zookeys.370.6291. ISSN 1313-2970. PMC 3904076. PMID 24478591.
- Geml J, Tulloss RE, Laursen GA, Sasanova NA, Taylor DL (2008). "Evidence for strong inter- and intracontinental phylogeographic structure in Amanita muscaria, a wind-dispersed ectomycorrhizal basidiomycete". Molecular Phylogenetics and Evolution. 48 (2): 694–701. doi:10.1016/j.ympev.2008.04.029. PMID 18547823.
- Roca AL, Georgiadis N, Pecon-Slattery J, O'Brien SJ (2001). "Genetic evidence for two species of elephant in Africa". Science. 293 (5534): 1473–1477. Bibcode:2001Sci...293.1473R. doi:10.1126/science.1059936. PMID 11520983. S2CID 38285623.
- Moran P, Kornfield I (1993). "Retention of an Ancestral Polymorphism in the Mbuna Species Flock (Teleostei: Cichlidae) of Lake Malawi" (PDF). Molecular Biology and Evolution. 10 (5): 1015–1029.
- Brown JK, Frohlich DR, Rosell RC (1995). "The sweetpotato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex?". Annual Review of Entomology. 40 (1): 511–534. doi:10.1146/annurev.en.40.010195.002455.
- De Queiroz, Kevin (2007). "Species Concepts and Species Delimitation". Systematic Biology. 56 (6): 879–886. doi:10.1080/10635150701701083. ISSN 1063-5157. PMID 18027281.
- Mayr E. (1970). "Morphological species characters and sibling species". Populations, Species, and Evolution. Cambridge, MA: The Belknap Press of Harvard University Press. pp. 21–36. ISBN 978-0-674-69013-4.
- Horandl, E.; Greilhuber, J.; Klimova, K.; Paun, O.; Temsch, E.; Emadzade, K.; Hodalova, I. (2009). "Reticulate evolution and taxonomic concepts in the Ranunculus auricomus complex (Ranunculaceae): insights from analysis of morphological, karyological and molecular data". Taxon. 58 (4): 1194–1215. doi:10.1002/tax.584012. PMC 2855680. PMID 20401184.
- JONG, R. (December 1987). "Superspecies Pyrgus malvae (Lepidoptera: Hesperiidae) in the East Mediterranean, with notes on phylogenetic and biological relationships". Rijksmuseum van Natuurlijke Historie – Via Naturalis Repository.
- Moritz C, Schneider CJ, Wake DB (1992). "Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation". Systematic Biology. 41 (3): 273–291. doi:10.1093/sysbio/41.3.273. S2CID 49267121.
- Bowen BW, Bass AL, Rocha LA, Grant WS, Roberston DR (2001). "Phylogeography of the trumpetfishes (Aulostomus): Ring species complex on a global scale". Evolution. 55 (5): 1029–1039. doi:10.1111/j.0014-3820.2001.tb00619.x. PMID 11430639. S2CID 221735739.
- Muñoz MM; Crawford NG; McGreevy Jr. TJ; Messana NJ; Tarvin RD; Revell LJ; Zandvliet RM; Hopwood JM; Mock E; Schneider AL; Schneider CJ. (2013). "Divergence in coloration and ecological speciation in the Anolis marmoratus species complex" (PDF). Molecular Ecology. 22 (10): 2668–2682. doi:10.1111/mec.12295. PMID 23611648. S2CID 16832446.
- Ride WD, Cogger HG, Dupuis C, Kraus O, Minelli A, Thompson FC, Tubbs PK, eds. (1999). "Chapter 9: Genus-group nominal taxa and their names". International Code of Zoological Nomenclature. London: The International Trust for Zoological Nomenclature. ISBN 978-0853010067.
- Lapage SP, Sneath PH, Lessel EF, Skerman VB, Seeliger HP, Clark WA, eds. (1992). "Chapter 3. Rules of nomenclature with recommendations. Section 2. Ranks of taxa". International Code of Nomenclature of Bacteria. Bacteriological Code, 1990 Revision. Washington (DC): ASM Press. ISBN 9781555810399.
- McNeill, J; et al., eds. (2012), International Code of Nomenclature for algae, fungi, and plants (Melbourne Code), adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011 (electronic ed.), Bratislava: International Association for Plant Taxonomy, retrieved 2012-12-20.
- Morris, Christopher, ed. (1992). "Physiologic race". Academic Press Dictionary of Science and Technology. San Diego / London: Academic Press. p. 1643. ISBN 978-0-12-200400-1.
- Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, Ingram KK, Das I (2007). "Cryptic species as a window on diversity and conservation" (PDF). Trends in Ecology & Evolution. 22 (3): 148–155. doi:10.1016/j.tree.2006.11.004. PMID 17129636.
- Mayr, Ernst (1942). Systematics and the Origin of Species, from the Viewpoint of a Zoologist. ISBN 9780674862500.
- Steyskal GC. (1972). "The meaning of the term 'sibling species'" (PDF). Systematic Zoology. 21 (4): 446. doi:10.1093/sysbio/21.4.446. Archived from the original (PDF) on 2015-02-07.
- Hodges SA, Arnold ML (1994). "Columbines: a geographically widespread species flock" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 91 (11): 5129–5132. Bibcode:1994PNAS...91.5129H. doi:10.1073/pnas.91.11.5129. PMC 43945. PMID 8197196.
- Fontdevila A, Pla C, Hasson E, Wasserman M, Sanchez A, Naveira H, Ruiz A (1988). "Drosophila koepferae: a new member of the Drosophila serido (Diptera: Drosophilidae) superspecies taxon". Annals of the Entomological Society of America. 81 (3): 380–385. doi:10.1093/aesa/81.3.380.
- Wallis GP, Arntzen JW (1989). "Mitochondrial-DNA variation in the crested newt superspecies: Limited cytoplasmic gene flow among species". Evolution. 43 (1): 88–104. doi:10.2307/2409166. JSTOR 2409166. PMID 28568488.
- Amadon D. (1966). "The superspecies concept". Systematic Biology. 15 (3): 245–249. doi:10.2307/sysbio/15.3.245.
- Heywood VH. (1962). "The "species aggregate" in theory and practice". In VH Heywood; Löve Á. (eds.). Symposium on Biosystematics, organized by the International Organization of Biosystematists, Montreal, October 1962. pp. 26–36.
- Kankare M, Van Nouhuys S, Hanski I (2005). "Genetic divergence among host-specific cryptic species in Cotesia melitaearum aggregate (Hymenoptera: Braconidae), parasitoids of checkerspot butterflies". Annals of the Entomological Society of America. 98 (3): 382–394. doi:10.1603/0013-8746(2005)098[0382:GDAHCS]2.0.CO;2.
- Wallis GP, Judge KF, Bland J, Waters JM, Berra TM (2001). "Genetic diversity in New Zealand Galaxias vulgaris sensu lato (Teleostei: Osmeriformes: Galaxiidae): a test of a biogeographic hypothesis". Journal of Biogeography. 28 (1): 59–67. doi:10.1046/j.1365-2699.2001.00535.x.
- Dai Y-C, Vainio EJ, Hantula J, Niemelä, Korhonen K (2003). "Investigations on Heterobasidion annosum s.lat. in central and eastern Asia with the aid of mating tests and DNA fingerprinting". Forest Pathology. 33 (5): 269–286. doi:10.1046/j.1439-0329.2003.00328.x.
- Van de Putte K, Nuytinck J, Stubbe D, Le HT, Verbeken A (2010). "Lactarius volemus sensu lato (Russulales) from northern Thailand: Morphological and phylogenetic species concepts explored". Fungal Diversity. 45 (1): 99–130. doi:10.1007/s13225-010-0070-0. S2CID 25615396.
- Tietze DT, Martens J, Sun YH (2006). "Molecular phylogeny of treecreepers (Certhia) detects hidden diversity". Ibis. 148 (3): 477–488. doi:10.1111/j.1474-919X.2006.00547.x.

- Marques, Isabel; Montgomery, Sean A.; Barker, Michael S.; Macfarlane, Terry D.; Conran, John G.; Catalán, Pilar; Rieseberg, Loren H.; Rudall, Paula J.; Graham, Sean W. (2016-04-01). "Transcriptome-derived evidence supports recent polyploidization and a major phylogeographic division in Trithuria submersa (Hydatellaceae, Nymphaeales)". New Phytologist. 210 (1): 310–323. doi:10.1111/nph.13755. ISSN 1469-8137. PMID 26612464.
- Steinfartz S, Veith M, Tautz D (2000). "Mitochondrial sequence analysis of Salamandra taxa suggests old splits of major lineages and postglacial recolonizations of Central Europe from distinct source populations of Salamandra salamandra". Molecular Ecology. 9 (4): 397–410. doi:10.1046/j.1365-294x.2000.00870.x. PMID 10736043. S2CID 10952136.
- Elmer K, Davila J, Lougheed S (2007). "Cryptic diversity and deep divergence in an upper Amazonian leaflitter frog, Eleutherodactylus ockendeni". BMC Evolutionary Biology. 7 (1): 247. doi:10.1186/1471-2148-7-247. ISSN 1471-2148. PMC 2254618. PMID 18154647.
- Mallet J, Beltrán M, Neukirchen W, Linares M (2007). "Natural hybridization in heliconiine butterflies: the species boundary as a continuum". BMC Evolutionary Biology. 7 (1): 28. doi:10.1186/1471-2148-7-28. PMC 1821009. PMID 17319954.

- "Iowa State University Department of Agronomy". Archived from the original on 2011-10-05. Retrieved 2015-01-01.
- "Species Group | Encyclopedia.com". www.encyclopedia.com. Retrieved 18 October 2019.
- (in Danish) Engelke, S. (2006?): Til Snegleforeningen (Note to the Danish Slug-society). Article in Danish
- Knowlton N (1993). "Sibling species in the sea". Annual Review of Ecology and Systematics. 24 (1): 189–216. doi:10.1146/annurev.es.24.110193.001201. ISSN 0066-4162.
- Knowlton N. (February 2000). "Molecular genetic analyses of species boundaries in the sea". Hydrobiologia. 420 (1): 73–90. doi:10.1023/A:1003933603879. ISSN 0018-8158. S2CID 40909997.
- Gómez A, Wright PJ, Lunt DH, Cancino JM, Carvalho GR, Hughes RN (2007). "Mating trials validate the use of DNA barcoding to reveal cryptic speciation of a marine bryozoan taxon". Proceedings of the Royal Society B. 274 (1607): 199–207. doi:10.1098/rspb.2006.3718. ISSN 0962-8452. PMC 1685843. PMID 17035167.
- Besansky NJ, Powell JR, Caccone A, Hamm DM, Scott JA, Collins FH (1994). "Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors". Proceedings of the National Academy of Sciences of the United States of America. 91 (15): 6885–8. Bibcode:1994PNAS...91.6885B. doi:10.1073/pnas.91.15.6885. PMC 44302. PMID 8041714.
- Clarke, A.R.; Powell, K.S.; Weldon, C.W.; Taylor, P.W. (2010-11-02). "The ecology of Bactrocera tryoni (Diptera: Tephritidae): what do we know to assist pest management?" (PDF). Annals of Applied Biology. 158 (1): 26–54. doi:10.1111/j.1744-7348.2010.00448.x. ISSN 0003-4746.
- Brown D, Brenneman R, Koepfli KP, Pollinger J, Mila B, Georgiadis N, Louis E, Grether G, Jacobs D, Wayne R (2007). "Extensive population genetic structure in the giraffe". BMC Biology. 5 (1): 57. doi:10.1186/1741-7007-5-57. ISSN 1741-7007. PMC 2254591. PMID 18154651.






.jpg.webp)
