TMEM126B
Transmembrane protein 126B is a protein that in humans is encoded by the TMEM126B gene.[4][5] TMEM126B is a mitochondrial transmembrane protein which is a component of the mitochondrial complex I assembly complex. The TMEM126B gene is conserved in mammals.[6] The encoded protein serves as an assembly factor that is required for formation of the membrane arm of the complex. It interacts with NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 13. Naturally occurring mutations in this gene are associated with isolated complex I deficiency. A pseudogene of this gene has been defined on chromosome 9.[4]
| TMEM126B | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||
| Aliases | TMEM126B, HT007, transmembrane protein 126B | ||||||||||||||||||||||||
| External IDs | OMIM: 615533 MGI: 1915722 HomoloGene: 10222 GeneCards: TMEM126B | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez | |||||||||||||||||||||||||
| Ensembl | |||||||||||||||||||||||||
| UniProt | |||||||||||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||||||||||
| RefSeq (protein) |
| ||||||||||||||||||||||||
| Location (UCSC) | n/a | Chr 7: 90.47 – 90.48 Mb | |||||||||||||||||||||||
| PubMed search | [2] | [3] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Structure
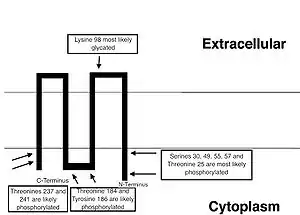
TMEM126B is located on the q arm of chromosome 11 in position 14.1 and has 7 exons.[4] The TMEM126B gene produces a 4.6 kDa protein composed of 54 amino acids.[7][8] It is a part of the mitochondrial complex I assembly (MCIA) complex, composed of NDUFAF1, ECSIT, and ACAD9 (by similarity). It associates with the intermediate 370 kDa subcomplex of incompletely assembled complex I.[9] Complex I is composed of 45 evolutionally conserved core subunits, including both mitochondrial DNA and nuclear encoded subunits. One of its arms is embedded in the inner membrane of the mitochondria, and the other is embedded in the organelle. The two arms are arranged in an L-shaped configuration. The total molecular weight of the complex is 1MDa.[10] A cartoon representation of the predicted orientation of TMEM126B within cell membrane, tentatively based on the phosphorylation[11] and hydrophobicity data[12] is shown below.
Function
The TMEM126B gene encodes a mitochondrial transmembrane protein which is a component of the mitochondrial complex I assembly complex. The encoded protein serves as an assembly factor that is required for formation of the membrane arm of the complex.[4] TMEM126B comigrates with other assembly factors including ACAD9, CIA30, and ECSIT. In the absence of TMEM126B, such assembly factors were not recruited into the mitochondrial membrane, and did not participate in complex I assembly. Dysfunction of TMEM126B has known to cause several complications in complex I characterized by severe difficulties in mitochondrial respiration and the complete failure of complex I assembly. However, it is not known to have significant effect on the assemblies of mitochondrial complexes III, IV, and V.[13]
Clinical Significance
Mutations in TMEM126B is known to result in mitochondrial diseases and associated disorders. It is majorly associated with a complex I deficiency, a deficiency in the first complex of the mitochondrial respiratory chain.[4] A complex I deficiency involving the dysfunction of the mitochondrial respiratory chain may cause a wide range of clinical manifestations from lethal neonatal disease to adult-onset neurodegenerative disorders. Phenotypes include macrocephaly with progressive leukodystrophy, non-specific encephalopathy, cardiomyopathy, Leigh syndrome, myopathy, liver disease, Leber hereditary optic neuropathy, and some forms of Parkinson disease.[9] In addition to complex I deficiency, TMEM126B mutations also show association with severe multi-system disorders during infancy, such as chronic renal failure and cardiomyopathy, and myopathy in childhood or adulthood.[13]
Discovery
TMEM126B was first discovered in a study of protein expression in tissues of the hypothalamus-pituitary-adrenal axis using full cDNA cloning.[5] It has since been detected in other tissues.[14]
Gene
TMEM126B is located on chromosome 11 in humans, flanked by the following genes:[15]
- DLG2: A member of the membrane-associated guanylate kinase family.
- TMEM126A: A paralog of TMEM126B expressed in the mitochondria.[16]
- CREBZF: Also known as the Zhangfei protein, a protein that interacts with herpes simplex virus.[17]
Translation
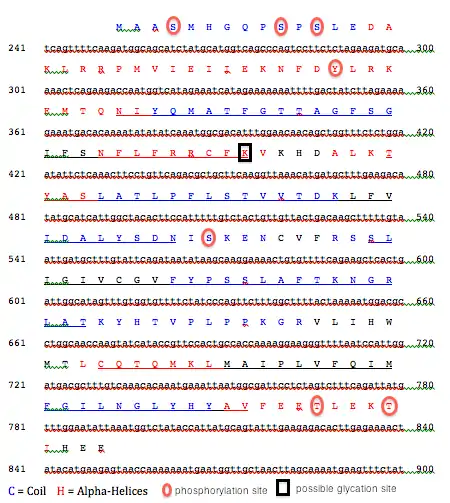
A conceptual translation of the TMEM126B protein, including a projection of the secondary structure,[18] predictions of transmembrane regions,[12] and putative phosphorylation[11] and glycation sites[19] is included below:
Tissue distribution
TMEM126B is expressed in most tissue types, with the notable exceptions of adipose tissue, ear tissue, the larynx, lymph tissue, nerve tissue, pituitary gland, spleen, thymus, thyroid, trachea, and umbilical cord.[20] It also appears to be highly expressed in parathyroid, bone marrow, and urinary bladder tissue.[20] There is also evidence that one of the isoforms of TMEM126B is expressed in the cell membrane of memory B cells of the adaptive immune system.[21]
Predicted properties
The following properties of TMEM126B were predicted using bioinformatic analysis:
- Molecular Weight: 22.7 KDal [22]
- Isoelectric point: 9.02[23]
- TMEM126B orthologs have highly variable isoelectric points.[23]
- Post-translational modification: several possible phosphorylation sites were detected,[11] and a pair of possible glycation sites were predicted[19]
- No signal peptide has been predicted.[24]
Interactions
In addition to co-subunits for complex I, TMEM126B has protein-protein interactions with ECSIT, NDUFAF1, NDUFC2, NDUFA13, and others.[9]
References
- GRCm38: Ensembl release 89: ENSMUSG00000030614 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: transmembrane protein 126B".
- Hu RM, Han ZG, Song HD, Peng YD, Huang QH, Ren SX, et al. (August 2000). "Gene expression profiling in the human hypothalamus-pituitary-adrenal axis and full-length cDNA cloning". Proceedings of the National Academy of Sciences of the United States of America. 97 (17): 9543–8. Bibcode:2000PNAS...97.9543H. doi:10.1073/pnas.160270997. PMC 16901. PMID 10931946.
- "HomoloGene Results: TMEM126B transmembrane protein 126B".
- Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, et al. (October 2013). "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research. 113 (9): 1043–53. doi:10.1161/CIRCRESAHA.113.301151. PMC 4076475. PMID 23965338.
- Yao, Daniel. "Cardiac Organellar Protein Atlas Knowledgebase (COPaKB) —— Protein Information". amino.heartproteome.org. Retrieved 2018-07-27.
- "Complex I assembly factor TMEM126B, mitochondrial". www.uniprot.org. Retrieved 2018-07-27.
- Rhein VF, Carroll J, Ding S, Fearnley IM, Walker JE (July 2016). "NDUFAF5 Hydroxylates NDUFS7 at an Early Stage in the Assembly of Human Complex I". The Journal of Biological Chemistry. 291 (28): 14851–60. doi:10.1074/jbc.M116.734970. PMC 4938201. PMID 27226634.
- "NetPhos 2.0 predictions for TMEM126B".
- Persson B, Argos P (March 1994). "Prediction of transmembrane segments in proteins utilising multiple sequence alignments". Journal of Molecular Biology. 237 (2): 182–92. doi:10.1006/jmbi.1994.1220. PMID 8126732.
- Alston CL, Compton AG, Formosa LE, Strecker V, Oláhová M, Haack TB, et al. (July 2016). "Biallelic Mutations in TMEM126B Cause Severe Complex I Deficiency with a Variable Clinical Phenotype". American Journal of Human Genetics. 99 (1): 217–27. doi:10.1016/j.ajhg.2016.05.021. PMC 5005451. PMID 27374774.
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. (December 2002). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proceedings of the National Academy of Sciences of the United States of America. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- "TMEM126B in Map Viewer".
- "Entrez Gene: TMEM126A transmembrane protein 126A".
- "OMIM Zhangfei protein".
- Burgess AW, Ponnuswamy PK, Sheraga HA (1974). "Analysis of conformations of amino acid residues and prediction of backbone topography in proteins". Israel Journal of Chemistry. 12 (1–2): 239–286. doi:10.1002/ijch.197400022.
- "NetGlycate 1.0 predictions for TMEM126B".
- "EST profile - Hs.525063".
- "CELLS BELONGING TO THE ADAPTIVE IMMUNE SYSTEM THAT EXPRESS PAQ ISOFORM OF TMEM126B".
- Brendel V, Bucher P, Nourbakhsh IR, Blaisdell BE, Karlin S (March 1992). "Methods and algorithms for statistical analysis of protein sequences". Proceedings of the National Academy of Sciences of the United States of America. 89 (6): 2002–6. Bibcode:1992PNAS...89.2002B. doi:10.1073/pnas.89.6.2002. PMC 48584. PMID 1549558.
- "PI Program (Isoelectric Point Prediction)". Archived from the original on 2008-10-26.
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (July 2004). "Improved prediction of signal peptides: SignalP 3.0". Journal of Molecular Biology. 340 (4): 783–95. CiteSeerX 10.1.1.165.2784. doi:10.1016/j.jmb.2004.05.028. PMID 15223320.
Further reading
- Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, Karasik D (September 2007). "Genome-wide association with bone mass and geometry in the Framingham Heart Study". BMC Medical Genetics. 8 Suppl 1: S14. doi:10.1186/1471-2350-8-S1-S14. PMC 1995606. PMID 17903296.
- Estrada K, Krawczak M, Schreiber S, van Duijn K, Stolk L, van Meurs JB, Liu F, Penninx BW, Smit JH, Vogelzangs N, Hottenga JJ, Willemsen G, de Geus EJ, Lorentzon M, von Eller-Eberstein H, Lips P, Schoor N, Pop V, de Keijzer J, Hofman A, Aulchenko YS, Oostra BA, Ohlsson C, Boomsma DI, Uitterlinden AG, van Duijn CM, Rivadeneira F, Kayser M (September 2009). "A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation". Human Molecular Genetics. 18 (18): 3516–24. doi:10.1093/hmg/ddp296. PMC 2729669. PMID 19570815.


