Cutinase
A cutinase (EC 3.1.1.74) is an enzyme that catalyzes the chemical reaction
- cutin + H2O cutin monomers
| Cutinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Cutinase | ||||||||
| Pfam | PF01083 | ||||||||
| InterPro | IPR000675 | ||||||||
| PROSITE | PDOC00140 | ||||||||
| SCOP2 | 1cex / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 127 | ||||||||
| OPM protein | 1oxm | ||||||||
| |||||||||
Thus, the two substrates of this enzyme are cutin and H2O, whereas its product is cutin monomer.
This enzyme belongs to the family of hydrolases, specifically those acting on carboxylic ester bonds. The systematic name of this enzyme class is cutin hydrolase.
Aerial plant organs are protected by a cuticle composed of an insoluble polymeric structural compound, cutin, which is a polyester composed of hydroxy and hydroxyepoxy fatty acids.[2] Plant pathogenic fungi produce extracellular degradative enzymes[3] that play an important role in pathogenesis. They include cutinase, which hydrolyses cutin, facilitating fungus penetration through the cuticle. Inhibition of the enzyme can prevent fungal infection through intact cuticles. Cutin monomers released from the cuticle by small amounts of cutinase on fungal spore surfaces can greatly increase the amount of cutinase secreted by the spore, the mechanism for which is as yet unknown.[2][3]
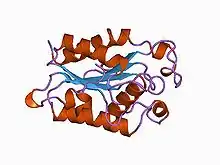
Cutinase is a serine esterase containing the classical Ser, His, Asp triad of serine hydrolases.[2] The protein belongs to the alpha-beta class, with a central beta-sheet of 5 parallel strands covered by 5 helices on either side of the sheet. The active site cleft is partly covered by 2 thin bridges formed by amino acid side chains, by contrast with the hydrophobic lid possessed by other lipases.[4] The protein also contains 2 disulfide bridges, which are essential for activity, their cleavage resulting in complete loss of enzymatic activity.[2] Two cutinase-like proteins (MtCY39.35 and MtCY339.08c) have been found in the genome of the bacteria Mycobacterium tuberculosis.
References
- Longhi S, Czjzek M, Lamzin V, Nicolas A, Cambillau C (May 1997). "Atomic resolution (1.0 A) crystal structure of Fusarium solani cutinase: stereochemical analysis". J. Mol. Biol. 268 (4): 779–99. doi:10.1006/jmbi.1997.1000. PMID 9175860.
- Ettinger WF, Thukral SK, Kolattukudy PE (1987). "Structure of cutinase gene, cDNA, and the derived amino acid sequence from phytopathogenic fungi". Biochemistry. 26 (24): 7883–7892. doi:10.1021/bi00398a052.
- Sweigard JA, Chumley FG, Valent B (1992). "Cloning and analysis of CUT1, a cutinase gene from Magnaporthe grisea". Mol. Gen. Genet. 232 (2): 174–182. doi:10.1007/BF00279994. PMID 1557023. S2CID 37444.
- Cambillau C, Martinez C, De Geus P, Lauwereys M, Matthyssens G (1992). "Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent". Nature. 356 (6370): 615–618. doi:10.1038/356615a0. PMID 1560844. S2CID 4334360.
- Garcia-Lepe R, Nuero OM, Reyes F, Santamaria F (1997). "Lipases in autolysed cultures of filamentous fungi". Lett. Appl. Microbiol. 25 (2): 127–30. doi:10.1046/j.1472-765X.1997.00187.x. PMID 9281862.
- Purdy RE, Kolattukudy PE (1975). "Hydrolysis of plant cuticle by plant pathogens. Purification, amino acid composition, and molecular weight of two isozymes of cutinase and a nonspecific esterase from Fusarium solani f. pisi". Biochemistry. 14 (13): 2824–31. doi:10.1021/bi00684a006. PMID 1156575.
- Purdy RE, Kolattukudy PE (1975). "Hydrolysis of plant cuticle by plant pathogens. Properties of cutinase I, cutinase II, and a nonspecific esterase isolated from Fusarium solani pisi". Biochemistry. 14 (13): 2832–40. doi:10.1021/bi00684a007. PMID 239740.
See also
| Wikimedia Commons has media related to Cutinase. |
| Look up cutinase in Wiktionary, the free dictionary. |