Acetylcholinesterase
Acetylcholinesterase (HGNC symbol ACHE; EC 3.1.1.7), also known as AChE or acetylhydrolase, is the primary cholinesterase in the body. It is an enzyme that catalyzes the breakdown of acetylcholine and of some other choline esters that function as neurotransmitters. AChE is found at mainly neuromuscular junctions and in chemical synapses of the cholinergic type, where its activity serves to terminate synaptic transmission. It belongs to carboxylesterase family of enzymes. It is the primary target of inhibition by organophosphorus compounds such as nerve agents and pesticides.
| acetylcholinesterase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
Acetylcholinesterase catalyzes the hydrolysis of acetylcholine to acetate ion and choline | |||||||||
| Identifiers | |||||||||
| EC number | 3.1.1.7 | ||||||||
| CAS number | 9000-81-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Enzyme structure and mechanism
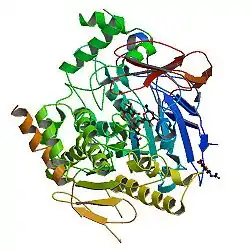
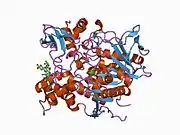
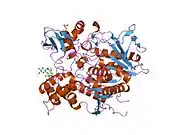
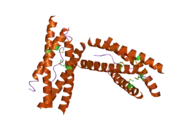
AChE is a hydrolase that hydrolyzes choline esters. It has a very high catalytic activity—each molecule of AChE degrades about 25,000 molecules of acetylcholine (ACh) per second, approaching the limit allowed by diffusion of the substrate.[6][7] The active site of AChE comprises 2 subsites—the anionic site and the esteratic subsite. The structure and mechanism of action of AChE have been elucidated from the crystal structure of the enzyme.[8][9]
The anionic subsite accommodates the positive quaternary amine of acetylcholine as well as other cationic substrates and inhibitors. The cationic substrates are not bound by a negatively charged amino acid in the anionic site, but by interaction of 14 aromatic residues that line the gorge leading to the active site.[10][11][12] All 14 amino acids in the aromatic gorge are highly conserved across different species.[13] Among the aromatic amino acids, tryptophan 84 is critical and its substitution with alanine results in a 3000-fold decrease in reactivity.[14] The gorge penetrates halfway through the enzyme and is approximately 20 angstroms long. The active site is located 4 angstroms from the bottom of the molecule.[15]
The esteratic subsite, where acetylcholine is hydrolyzed to acetate and choline, contains the catalytic triad of three amino acids: serine 200, histidine 440 and glutamate 327. These three amino acids are similar to the triad in other serine proteases except that the glutamate is the third member rather than aspartate. Moreover, the triad is of opposite chirality to that of other proteases.[16] The hydrolysis reaction of the carboxyl ester leads to the formation of an acyl-enzyme and free choline. Then, the acyl-enzyme undergoes nucleophilic attack by a water molecule, assisted by the histidine 440 group, liberating acetic acid and regenerating the free enzyme.[17][18]
Biological function
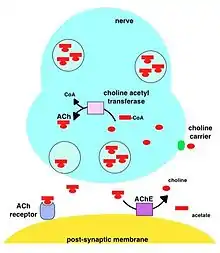
During neurotransmission, ACh is released from the presynaptic neuron into the synaptic cleft and binds to ACh receptors on the post-synaptic membrane, relaying the signal from the nerve. AChE, also located on the post-synaptic membrane, terminates the signal transmission by hydrolyzing ACh. The liberated choline is taken up again by the pre-synaptic neuron and ACh is synthesized by combining with acetyl-CoA through the action of choline acetyltransferase.[19][20]
A cholinomimetic drug disrupts this process by acting as a cholinergic neurotransmitter that is impervious to acetylcholinesterase's lysing action.
Disease relevance
For a cholinergic neuron to receive another impulse, ACh must be released from the ACh receptor. This occurs only when the concentration of ACh in the synaptic cleft is very low. Inhibition of AChE leads to accumulation of ACh in the synaptic cleft and results in impeded neurotransmission.
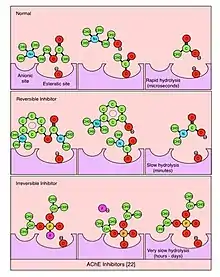
Irreversible inhibitors of AChE may lead to muscular paralysis, convulsions, bronchial constriction, and death by asphyxiation. Organophosphates (OP), esters of phosphoric acid, are a class of irreversible AChE inhibitors.[21] Cleavage of OP by AChE leaves a phosphoryl group in the esteratic site, which is slow to be hydrolyzed (on the order of days) and can become covalently bound. Irreversible AChE inhibitors have been used in insecticides (e.g., malathion) and nerve gases for chemical warfare (e.g., Sarin and Soman). Carbamates, esters of N-methyl carbamic acid, are AChE inhibitors that hydrolyze in hours and have been used for medical purposes (e.g., physostigmine for the treatment of glaucoma). Reversible inhibitors occupy the esteratic site for short periods of time (seconds to minutes) and are used to treat of a range of central nervous system diseases. Tetrahydroaminoacridine (THA) and donepezil are FDA-approved to improve cognitive function in Alzheimer's disease. Rivastigmine is also used to treat Alzheimer's and Lewy body dementia, and pyridostigmine bromide is used to treat myasthenia gravis.[22][23][24][25][26][27]
An endogenous inhibitor of AChE in neurons is Mir-132 microRNA, which may limit inflammation in the brain by silencing the expression of this protein and allowing ACh to act in an anti-inflammatory capacity.[28]
It has also been shown that the main active ingredient in cannabis, tetrahydrocannabinol, is a competitive inhibitor of acetylcholinesterase.[29]
Distribution
AChE is found in many types of conducting tissue: nerve and muscle, central and peripheral tissues, motor and sensory fibers, and cholinergic and noncholinergic fibers. The activity of AChE is higher in motor neurons than in sensory neurons.[30][31][32]
Acetylcholinesterase is also found on the red blood cell membranes, where different forms constitute the Yt blood group antigens.[33] Acetylcholinesterase exists in multiple molecular forms, which possess similar catalytic properties, but differ in their oligomeric assembly and mode of attachment to the cell surface.
AChE gene
In mammals, acetylcholinesterase is encoded by a single AChE gene while some invertebrates have multiple acetylcholinesterase genes. Note higher vertebrates also encode a closely related paralog BCHE (butyrylcholinesterase) with 50% amino acid identity to ACHE.[34] Diversity in the transcribed products from the sole mammalian gene arises from alternative mRNA splicing and post-translational associations of catalytic and structural subunits. There are three known forms: T (tail), R (read through), and H(hydrophobic).[35]
AChET
The major form of acetylcholinesterase found in brain, muscle, and other tissues, known as is the hydrophilic species, which forms disulfide-linked oligomers with collagenous, or lipid-containing structural subunits. In the neuromuscular junctions AChE expresses in asymmetric form which associates with ColQ or subunit. In the central nervous system it is associated with PRiMA which stands for Proline Rich Membrane anchor to form symmetric form. In either case, the ColQ or PRiMA anchor serves to maintain the enzyme in the intercellular junction, ColQ for the neuromuscular junction and PRiMA for synapses.
AChEH
The other, alternatively spliced form expressed primarily in the erythroid tissues, differs at the C-terminus, and contains a cleavable hydrophobic peptide with a PI-anchor site. It associates with membranes through the phosphoinositide (PI) moieties added post-translationally.[36]
AChER
The third type has, so far, only been found in Torpedo sp. and mice although it is hypothesized in other species. It is thought to be involved in the stress response and, possibly, inflammation.[37]
Nomenclature
The nomenclatural variations of ACHE and of cholinesterases generally are discussed at Cholinesterase § Types and nomenclature.
Inhibitors
For acetylcholine esterase (AChE), reversible inhibitors are those that do not irreversibly bond to and deactivate AChE.[38] Drugs that reversibly inhibit acetylcholine esterase are being explored as treatments for Alzheimer's disease and myasthenia gravis, among others. Examples include tacrine and donepezil.[39]
See also
 Biology portal
Biology portal- Acetylcholinesterase inhibitor
- Cholinesterases
References
- GRCh38: Ensembl release 89: ENSG00000087085 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000023328 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Katzung BG (2001). Basic and clinical pharmacology:Introduction to autonomic pharmacology (8 ed.). The McGraw Hill Companies. pp. 75–91. ISBN 978-0-07-160405-5.
- Quinn DM (1987). "Acetylcholinesterase: enzyme structure, reaction dynamics, and virtual transition states". Chemical Reviews. 87 (5): 955–79. doi:10.1021/cr00081a005.
- Taylor P, Radić Z (1994). "The cholinesterases: from genes to proteins". Annual Review of Pharmacology and Toxicology. 34: 281–320. doi:10.1146/annurev.pa.34.040194.001433. PMID 8042853.
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I (August 1991). "Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein". Science. 253 (5022): 872–9. Bibcode:1991Sci...253..872S. doi:10.1126/science.1678899. PMID 1678899. S2CID 28833513.
- Sussman JL, Harel M, Silman I (June 1993). "Three-dimensional structure of acetylcholinesterase and of its complexes with anticholinesterase drugs". Chem. Biol. Interact. 87 (1–3): 187–97. doi:10.1016/0009-2797(93)90042-W. PMID 8343975.
- Radić Z, Gibney G, Kawamoto S, MacPhee-Quigley K, Bongiorno C, Taylor P (October 1992). "Expression of recombinant acetylcholinesterase in a baculovirus system: kinetic properties of glutamate 199 mutants". Biochemistry. 31 (40): 9760–7. doi:10.1021/bi00155a032. PMID 1356436.
- Ordentlich A, Barak D, Kronman C, Ariel N, Segall Y, Velan B, Shafferman A (February 1995). "Contribution of aromatic moieties of tyrosine 133 and of the anionic subsite tryptophan 86 to catalytic efficiency and allosteric modulation of acetylcholinesterase". J. Biol. Chem. 270 (5): 2082–91. doi:10.1074/jbc.270.5.2082. PMID 7836436.
- Ariel N, Ordentlich A, Barak D, Bino T, Velan B, Shafferman A (October 1998). "The 'aromatic patch' of three proximal residues in the human acetylcholinesterase active centre allows for versatile interaction modes with inhibitors". Biochem. J. 335 (1): 95–102. doi:10.1042/bj3350095. PMC 1219756. PMID 9742217.

- Ordentlich A, Barak D, Kronman C, Flashner Y, Leitner M, Segall Y, Ariel N, Cohen S, Velan B, Shafferman A (August 1993). "Dissection of the human acetylcholinesterase active center determinants of substrate specificity. Identification of residues constituting the anionic site, the hydrophobic site, and the acyl pocket". J. Biol. Chem. 268 (23): 17083–95. PMID 8349597.

- Tougu V (2001). "Acetylcholinesterase: Mechanism of Catalysis and Inhibition". Current Medicinal Chemistry-Central Nervous System Agents. 1 (2): 155–170. doi:10.2174/1568015013358536.

- Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, Axelsen PH, Silman I, Sussman JL (1993). "Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase". Proceedings of the National Academy of Sciences of the United States of America. 90 (19): 9031–5. Bibcode:1993PNAS...90.9031H. doi:10.1073/pnas.90.19.9031. PMC 47495. PMID 8415649.
- Tripathi A (October 2008). "Acetylcholinsterase: A Versatile Enzyme of Nervous System". Annals of Neurosciences. 15 (4): 106–111. doi:10.5214/ans.0972.7531.2008.150403.
- Pauling L (1946). "Molecular Architecture and Biological Reactions" (PDF). Chemical & Engineering News. 24 (10): 1375–1377. doi:10.1021/cen-v024n010.p1375.
- Fersht A (1985). Enzyme structure and mechanism. San Francisco: W.H. Freeman. p. 14. ISBN 0-7167-1614-3.
- Whittaker VP (1990). "The Contribution of Drugs and Toxins to Understanding of Cholinergic Function" (PDF). Trends in Pharmacological Sciences. 11 (1): 8–13. doi:10.1016/0165-6147(90)90034-6. hdl:11858/00-001M-0000-0013-0E8C-5. PMID 2408211.
- Purves D, Augustine GJ, Fitzpatrick D, Hall WC, LaMantia AS, McNamara JO, White LE (2008). Neuroscience (4th ed.). Sinauer Associates. pp. 121–2. ISBN 978-0-87893-697-7.
- "National Pesticide Information Center-Diazinon Technical Fact Sheet" (PDF). Retrieved 24 February 2012.
- "Clinical Application: Acetylcholine and Alzheimer's Disease". Retrieved 24 February 2012.
- Stoelting RK (1999). Anticholinesterase Drugs and Cholinergic Agonists", in Pharmacology and Physiology in Anesthetic Practice. Lippincott-Raven. ISBN 978-0-7817-5469-9. Archived from the original on 2016-03-03. Retrieved 2012-02-26.
- Taylor P, Hardman JG, Limbird LE, Molinoff PB, Ruddon RW, Gilman AG (1996). "5: Autonomic Pharmacology: Cholinergic Drugs". The Pharmacologial Basis of Therapeutics. THe McGraw-Hill Companies. pp. 161–174. ISBN 978-0-07-146804-6.
- Blumenthal D, Brunton L, Goodman LS, Parker K, Gilman A, Lazo JS, Buxton I (1996). "5: Autonomic Pharmacology: Cholinergic Drugs". Goodman & Gilman's The pharmacological basis of therapeutics. New York: McGraw-Hill. p. 1634. ISBN 978-0-07-146804-6.
- Drachman DB, Isselbacher KJ, Braunwald E, Wilson JD, Martin JB, Fauci AS, Kasper DL (1998). Harrison's Principles of Internal Medicine (14 ed.). The McCraw-Hill Companies. pp. 2469–2472. ISBN 978-0-07-020291-7.
- Raffe RB (2004). Autonomic and Somatic Nervous Systems in Netter's Illustrated Pharmacology. Elsevier Health Science. p. 43. ISBN 978-1-929007-60-8.
- Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H (2009). "MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase". Immunity. 31 (6): 965–73. doi:10.1016/j.immuni.2009.09.019. PMID 20005135.
- Eubanks LM, Rogers CJ, Beuscher AE, Koob GF, Olson AJ, Dickerson TJ, Janda KD (2006). "A molecular link between the active component of marijuana and Alzheimer's disease pathology". Mol. Pharm. 3 (6): 773–7. doi:10.1021/mp060066m. PMC 2562334. PMID 17140265.
- Massoulié J, Pezzementi L, Bon S, Krejci E, Vallette FM (July 1993). "Molecular and cellular biology of cholinesterases". Progress in Neurobiology. 41 (1): 31–91. doi:10.1016/0301-0082(93)90040-Y. PMID 8321908. S2CID 21601586.
- Chacko LW, Cerf JA (1960). "Histochemical localization of cholinesterase in the amphibian spinal cord and alterations following ventral root section". Journal of Anatomy. 94 (Pt 1): 74–81. PMC 1244416. PMID 13808985.
- Koelle GB (1954). "The histochemical localization of cholinesterases in the central nervous system of the rat". Journal of Comparative Anatomy. 100 (1): 211–35. doi:10.1002/cne.901000108. PMID 13130712. S2CID 23021010.
- Bartels CF, Zelinski T, Lockridge O (May 1993). "Mutation at codon 322 in the human acetylcholinesterase (ACHE) gene accounts for YT blood group polymorphism". Am. J. Hum. Genet. 52 (5): 928–36. PMC 1682033. PMID 8488842.
- Johnson G, Moore SW (2012). "Why has butyrylcholinesterase been retained? Structural and functional diversification in a duplicated gene. 2012". Neurochem. Int. 16 (5): 783–797. doi:10.1016/j.neuint.2012.06.016. PMID 22750491. S2CID 39348660.
- Massoulié J, Perrier N, Noureddine H, Liang D, Bon S (2008). "Old and new questions about cholinesterases". Chem. Biol. Interact. 175 (1–3): 30–44. doi:10.1016/j.cbi.2008.04.039. PMID 18541228.
- "Entrez Gene: ACHE acetylcholinesterase (Yt blood group)".
- Dori A, Ifergane G, Saar-Levy T, Bersudsky M, Mor I, Soreq H, Wirguin I (2007). "Readthrough acetylcholinesterase in inflammation-associated neuropathies". Life Sci. 80 (24–25): 2369–74. doi:10.1016/j.lfs.2007.02.011. PMID 17379257.
- Millard CB, Kryger G, Ordentlich A, Greenblatt HM, Harel M, Raves ML, Segall Y, Barak D, Shafferman A, Silman I, Sussman JL (June 1999). "Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level". Biochemistry. 38 (22): 7032–9. doi:10.1021/bi982678l. PMID 10353814.
- Julien RM, Advokat CD, Comaty JE (2007-10-12). A Primer of Drug Action (Eleventh ed.). Worth Publishers. pp. 50. ISBN 978-1-4292-0679-2.
Further reading
- Silman I, Futerman AH (1988). "Modes of attachment of acetylcholinesterase to the surface membrane". Eur. J. Biochem. 170 (1–2): 11–22. doi:10.1111/j.1432-1033.1987.tb13662.x. PMID 3319614.
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I (1991). "Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein". Science. 253 (5022): 872–9. Bibcode:1991Sci...253..872S. doi:10.1126/science.1678899. PMID 1678899. S2CID 28833513.
- Soreq H, Seidman S (2001). "Acetylcholinesterase--new roles for an old actor". Nature Reviews Neuroscience. 2 (4): 294–302. doi:10.1038/35067589. PMID 11283752. S2CID 5947744.
- Shen T, Tai K, Henchman RH, McCammon JA (2003). "Molecular dynamics of acetylcholinesterase". Acc. Chem. Res. 35 (6): 332–40. doi:10.1021/ar010025i. PMID 12069617.
- Pakaski M, Kasa P (2003). "Role of acetylcholinesterase inhibitors in the metabolism of amyloid precursor protein". Current Drug Targets. CNS and Neurological Disorders. 2 (3): 163–71. doi:10.2174/1568007033482869. PMID 12769797.
- Meshorer E, Soreq H (2006). "Virtues and woes of AChE alternative splicing in stress-related neuropathologies". Trends Neurosci. 29 (4): 216–24. doi:10.1016/j.tins.2006.02.005. PMID 16516310. S2CID 18983474.
- Ehrlich G, Viegas-Pequignot E, Ginzberg D, Sindel L, Soreq H, Zakut H (1992). "Mapping the human acetylcholinesterase gene to chromosome 7q22 by fluorescent in situ hybridization coupled with selective PCR amplification from a somatic hybrid cell panel and chromosome-sorted DNA libraries". Genomics. 13 (4): 1192–7. doi:10.1016/0888-7543(92)90037-S. PMID 1380483.
- Spring FA, Gardner B, Anstee DJ (1992). "Evidence that the antigens of the Yt blood group system are located on human erythrocyte acetylcholinesterase". Blood. 80 (8): 2136–41. doi:10.1182/blood.V80.8.2136.2136. PMID 1391965.
- Shafferman A, Kronman C, Flashner Y, Leitner M, Grosfeld H, Ordentlich A, Gozes Y, Cohen S, Ariel N, Barak D (1992). "Mutagenesis of human acetylcholinesterase. Identification of residues involved in catalytic activity and in polypeptide folding". J. Biol. Chem. 267 (25): 17640–8. PMID 1517212.
- Getman DK, Eubanks JH, Camp S, Evans GA, Taylor P (1992). "The human gene encoding acetylcholinesterase is located on the long arm of chromosome 7". Am. J. Hum. Genet. 51 (1): 170–7. PMC 1682883. PMID 1609795.
- Li Y, Camp S, Rachinsky TL, Getman D, Taylor P (1992). "Gene structure of mammalian acetylcholinesterase. Alternative exons dictate tissue-specific expression". J. Biol. Chem. 266 (34): 23083–90. PMID 1744105.
- Velan B, Grosfeld H, Kronman C, Leitner M, Gozes Y, Lazar A, Flashner Y, Marcus D, Cohen S, Shafferman A (1992). "The effect of elimination of intersubunit disulfide bonds on the activity, assembly, and secretion of recombinant human acetylcholinesterase. Expression of acetylcholinesterase Cys-580----Ala mutant". J. Biol. Chem. 266 (35): 23977–84. PMID 1748670.
- Soreq H, Ben-Aziz R, Prody CA, Seidman S, Gnatt A, Neville L, Lieman-Hurwitz J, Lev-Lehman E, Ginzberg D, Lipidot-Lifson Y (1991). "Molecular cloning and construction of the coding region for human acetylcholinesterase reveals a G + C-rich attenuating structure". Proceedings of the National Academy of Sciences of the United States of America. 87 (24): 9688–92. Bibcode:1990PNAS...87.9688S. doi:10.1073/pnas.87.24.9688. PMC 55238. PMID 2263619.
- Chhajlani V, Derr D, Earles B, Schmell E, August T (1989). "Purification and partial amino acid sequence analysis of human erythrocyte acetylcholinesterase". FEBS Lett. 247 (2): 279–82. doi:10.1016/0014-5793(89)81352-3. PMID 2714437. S2CID 41843002.
- Lapidot-Lifson Y, Prody CA, Ginzberg D, Meytes D, Zakut H, Soreq H (1989). "Coamplification of human acetylcholinesterase and butyrylcholinesterase genes in blood cells: correlation with various leukemias and abnormal megakaryocytopoiesis". Proceedings of the National Academy of Sciences of the United States of America. 86 (12): 4715–9. Bibcode:1989PNAS...86.4715L. doi:10.1073/pnas.86.12.4715. PMC 287342. PMID 2734315.
- Bazelyansky M, Robey E, Kirsch JF (1986). "Fractional diffusion-limited component of reactions catalyzed by acetylcholinesterase". Biochemistry. 25 (1): 125–30. doi:10.1021/bi00349a019. PMID 3954986.
- Gaston SM, Marchase RB, Jakoi ER (1982). "Brain ligatin: a membrane lectin that binds acetylcholinesterase". J. Cell. Biochem. 18 (4): 447–59. doi:10.1002/jcb.1982.240180406. PMID 7085778. S2CID 22975039.
- Ordentlich A, Barak D, Kronman C, Ariel N, Segall Y, Velan B, Shafferman A (1995). "Contribution of aromatic moieties of tyrosine 133 and of the anionic subsite tryptophan 86 to catalytic efficiency and allosteric modulation of acetylcholinesterase". J. Biol. Chem. 270 (5): 2082–91. doi:10.1074/jbc.270.5.2082. PMID 7836436.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Ben Aziz-Aloya R, Sternfeld M, Soreq H (1994). "Promoter elements and alternative splicing in the human ACHE gene". Prog. Brain Res. 98: 147–53. doi:10.1016/s0079-6123(08)62392-4. PMID 8248502.
- Massoulié J, Pezzementi L, Bon S, Krejci E, Vallette FM (1993). "Molecular and Cellular Biology of Cholinesterases". Prog. Brain Res. 41 (1): 31–91. doi:10.1016/0301-0082(93)90040-Y. PMID 8321908. S2CID 21601586.
External links
- ATSDR Case Studies in Environmental Medicine: Cholinesterase Inhibitors, Including Insecticides and Chemical Warfare Nerve Agents U.S. Department of Health and Human Services
- Proteopedia Acetylcholinesterase
- Proteopedia AChE_inhibitors_and_substrates
- Proteopedia AChE_inhibitors_and_substrates_(Part_II)
- Proteopedia AChE bivalent inhibitors AChE_bivalent_inhibitors AChE bivalent inhibitors
- Acetylcholinesterase: A gorge-ous enzyme—PDBe
- Acetylcholinesterase—RCSB PDB
- Human ACHE genome location and ACHE gene details page in the UCSC Genome Browser.
- Overview of all the structural information available in the PDB for UniProt: P22303 (Human Acetylcholinesterase) at the PDBe-KB.