Formylation
In biochemistry, the addition of a formyl functional group is termed formylation. A formyl functional group consists of a carbonyl bonded to hydrogen. When attached to an R group, a formyl group is called an aldehyde.

Formylation has been identified in several critical biological processes. Methionine was first discovered to be formylated in E. coli by Marcker and Sanger in 1964[1] and was later identified to be involved in the initiation of protein synthesis in bacteria and organelles.[2] The formation of N-formylmethionine is catalyzed by the enzyme methionyl-tRNAMet transformylase.[3] Additionally, two formylation reactions occur in the de novo biosynthesis of purines. These reactions are catalyzed by the enzymes glycinamide ribonucleotide (GAR) transformylase and 5-aminoimidazole-4-carboxyamide ribotide (AICAR) transformylase.[4] More recently, formylation has been discovered to be a histone modification, which may modulate gene expression.
General formylation reaction
Formylation refers to any chemical processes in which a compound is functionalized with a formyl group (-CH=O). In organic chemistry, the term is most commonly used with regards to aromatic compounds (for example the conversion of benzene to benzaldehyde in the Gattermann–Koch reaction). In biochemistry the reaction is catalysed by enzymes such as formyltransferases.
Formylation reactions in biology
Methanogenesis
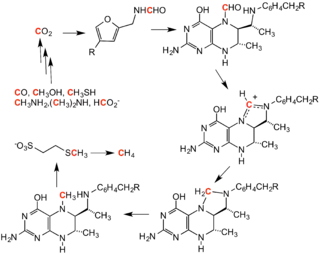
Formylation of methanofuran initiates the methanogenesis cycle. The formyl group is derived from carbon dioxide and is converted to methane.
Formylation in protein synthesis

In bacteria and organelles, the initiation of protein synthesis is signaled by the formation of formyl-methionyl-tRNA (tRNAfMet). This reaction is dependent on 10-formyltetrahydrofolate, and the enzyme methionyl-tRNA formyltransferase.[3] This reaction is not used by eukaryotes or Archaea, as the presence of tRNAfMet in non bacterial cells is dubbed as intrusive material and quickly eliminated. After its production, tRNAfMet is delivered to the 30S subunit of the ribosome in order to start protein synthesis. fMet possesses the same codon sequence as methionine. However, fMet is only used for the initiation of protein synthesis and is thus found only at the N terminus of the protein. Methionine is used during the rest translation. In E. coli, tRNAfMet is specifically recognized by initiation factor IF-2, as the formyl group blocks peptide bond formation at the N-terminus of methionine.[3]
Once protein synthesis is accomplished, the formyl group on methionine can be removed by peptide deformylase. The methionine residue can be further removed by the enzyme methionine aminopeptidase.

Formylation reactions in purine biosynthesis
Two formylation reactions are required in the eleven step de novo synthesis of inosine monophosphate (IMP), the precursor of the purine ribonucleotides AMP and GMP. Glycinamide ribonucleotide (GAR) transformylase catalyzes the formylation of GAR to formylglycinamidine ribotide (FGAR) in the fourth reaction of the pathway. In the penultimate step of de novo purine biosynthesis, 5-aminoimidazole-4-carboxyamide ribotide (AICAR) is formylated to 5-formaminoimidazole-4-carboxamide ribotide (FAICAR) by AICAR transformylase.[4]
GAR transformylase
PurN GAR transformylase is found in eukaryotes and prokaryotes. However, a second GAR transformylase, PurT GAR transformylase has been identified in E. coli. While the two enzymes have no sequence conservation and require different formyl donors, the specific activity and Km for GAR are the same in both PurT and PurN GAR transformylase.
PurN GAR transformylase
PurN GAR transformylase 1CDE uses the coenzyme N10-formyltetrahydrofolate (N10-formyl-THF) as a formyl donor to formylate the α-amino group of GAR. In eukaryotes, PurN GAR transformylase is part of a large multifunctional protein, but is found as a single protein in prokaryotes.[6]
Mechanism

The formylation reaction is proposed to occur through a direct transfer reaction in which the amine group of GAR nucleophillically attacks N10-formyl-THF creating a tetrahedral intermediate.[4] As the α-amino group of GAR is relatively reactive, deprotonation of the nucleophile is proposed to occur by solvent. In the active site, Asn 106, His 108, and Asp 144 are positioned to assist with formyl transfer.[6] However, mutagenesis studies have indicated that these residues are not individually essential for catalysis, as only mutations of two or more residues inhibit the enzyme. Based on the structure the negatively charged Asp144 is believed to increase the pKa of His108, allowing the protonated imidazolium group of His108 to enhances the electrophillicity of the N10-formyl-THF formyl group. Additionally, His108 and Asn106 are believed to stabilize the oxyanion formed in the transition state.[7]

PurT GAR transformylase
PurT GAR transformylase requires formate as the formyl donor and ATP for catalysis. It has been estimated that PurT GAR transformylase carries out 14-50% of GAR formylations in E. coli. The enzyme is a member of the ATP-grasp superfamily of proteins.[8]
Mechanism
A sequential mechanism has been proposed for PurT GAR transformylase in which a short lived formyl phosphate intermediate is proposed to first form. This formyl phosphate intermediate then undergoes nucleophillic attack by the GAR amine for transfer of the formyl group. A formyl phosphate intermediate has been detected in mutagenesis experiments, in which the mutant PurT GAR transforymylase had a weak affinity for formate.[6] Incubating PurT GAR transformylase with formyl phosphate, ADP, and GAR, yields both ATP and FGAR. This further indicating that formyl phosphate may be an intermediate, as it is kinetically and chemically competent to carry out the formylation reaction in the enzyme.[9] A enzyme phosphate intermediate preceding the formylphosphate intermediate has also been proposed to form based on positional isotope exchange studies.[9] However, structural data indicates that the formate may be positioned for a direct attack on the γ-phosphate of ATP in the enzyme’s active site to form the formylphosphate intermediate.[8]

AICAR transformylase
AICAR transformylase requires the coenzyme N10-formyltetrahydrofolate (N10-formyl-THF) as the formyl donor for the formylation of AICAR to FAICAR. However, AICAR transformylase and GAR transformylase do not share a high sequence similarity or structural homology.[7]
Mechanism
The amine on AICAR is much less nucleophillic than its counterpart on GAR due to delocalization of electrons in AICAR through conjugation. Therefore, the N5 nucleophile of AIRCAR must be activated for the formylation reaction to occur. Histidine 268 and Lysine 267 have been found to be essential for catalysis and are conserved in all AICAR transformylase. Histidine 268 is involved in deprotonation of the N5 nucleophile of AICAR, whereas Lysine 267 is proposed to stabilize the tetrahedral intermediate.[7]

Formylation in Histone Proteins

ε-Formylation is one of many post-translational modifications that occur on histone proteins, which been shown to modulate chromatin conformations and gene activation.

Formylation has been identified on the Nε of lysine residues in histones and proteins. This modification has been observed in linker histones and high mobility group proteins, it is highly abundant and it is believed to have a role in the epigenetics of chromatin function. Lysines that are formylated have been shown to play a role in DNA binding. Additionally, formylation has been detected on histone lysines that are also known to be acetylated and methylated. Thus, formylation may block other post-translational modifications.[10] Formylation is detected most frequently on 19 different modification sites on Histone H1. The genetic expression of the cell is highly disrupted by formylation, which may cause diseases such as cancer. The development of these modifications may be due to oxidative stress.[10]
In histone proteins, lysine is typically modified by Histone Acetyl-Transferases (HATs) and Histone Deacetylases (HDAC or KDAC). The acetylation of lysine is fundamental to the regulation and expression of certain genes. Oxidative stress creates a significantly different environment in which acetyl-lysine can be quickly outcompeted by the formation of formyl-lysine due to the high reactivity of formylphosphate species. This situation is currently believed to be caused by oxidative DNA damage. A mechanism for the formation of formylphosphate has been proposed, which it is highly dependent on oxidatively damaged DNA and mainly driven by radical chemistry within the cell.[11] The formylphosphate produced can then be used to formylate lysine. Oxidative stress is believed to play a role in the availability of lysine residues in the surface of proteins and the possibility of being formylated.

Formylation in medicine
Formylation reactions as a drug target
Inhibition of enzymes involved in purine biosynthesis has been exploited as a potential drug target for chemotherapy.

Cancer cells require high concentrations of purines to facilitate division[6] and tend to rely on de novo synthesis rather than the nucleotide salvage pathway.[7][12] Several folate based inhibitors have been developed to inhibit formylation reactions by GAR transformylase and AICAR transformylase.[13] The first GAR transformylase inhibitor Lometrexol [(6R)5,10-dideazatetrahydrofolate] was developed in the 1980s through a collaboration between Eli Lilly and academic laboratories.[14]
Although similar in structure to N10-formyl-THF, lometrexol is incapable of carrying out one carbon transfer reactions.[13] Additionally, several GAR based inhibitors of GAR transformylase have also been synthesized.[13] Development of folate based inhibitors have been found to be particularly challenging as the inhibitors also down regulate the enzyme folypolyglutamate synthase, which adds additional γ-glutamates to monoglutamate folates and antifolates after entering the cell for increased enzyme affinity. This increased affinity can lead to antifolate resistance.[12]
Leigh Syndrome
Leigh syndrome is a neurodegenerative disorder that has been linked to a defect in an enzymatic formylation reaction. Leigh syndrome is typically associated with defects in oxidative phosphorylation, which occurs in the mitochondria.[15] Exome sequencing, has been used to identify a mutation in the gene coding for mitochondrial methionyl-tRNA formyltransferase (MTFMT) in patients with Leigh syndrome. The c.626C>T mutation identified in MTFMT yielding symptoms of Leigh Syndrome is believed to alter exon splicing leading to a frameshift mutation and a premature stop codon. Individuals with the MTFMT c.626C>T mutation were found to have reduced fMet-tRNAMet levels and changes in the formylation level of mitochondrically translated COX1. This link provides evidence for the necessity of formylated methionine in initiation of expression for certain mitochondrial genes.[16]
References
- Marcker, K; Sanger, F. (1964). "N-formyl-methionyl-S-RNA". J. Mol. Biol. 8 (6): 835–840. doi:10.1016/S0022-2836(64)80164-9. PMID 14187409.
- Adams, J.M.; Capecchi, M.R. (1966). "N-Formylmethionyl-sRNA as the initiator of protein synthesis". PNAS. 55 (1): 147–155. Bibcode:1966PNAS...55..147A. doi:10.1073/pnas.55.1.147. PMC 285768. PMID 5328638.
- Kozak, M (1983). "Comparison of Initiation of Protein synthesis in Procaryotes, Eucaryotes, and Organelles". Microbiological Reviews. 47 (1): 1–45. doi:10.1128/MMBR.47.1.1-45.1983. PMC 281560. PMID 6343825.
- Voet and Voet (2008). Fundamentals of Biochemistry 3rd edition. New York: Wiley.
- Thauer, R. K. (1998). "Biochemistry of Methanogenesis: a Tribute to Marjory Stephenson". Microbiology. 144: 2377–2406. doi:10.1099/00221287-144-9-2377. PMID 9782487.
- Warren, M.S.; K.M. Mattia; A.E. Marolewski; S.J. Benkovic (1996). "The transformylase enzymes of de novo purine biosynthesis" (PDF). Pure Appl. Chem. 68 (11): 2029–2036. doi:10.1351/pac199668112029. S2CID 39555269. Retrieved 24 February 2013.
- Wolan, D; Greasley, S.E.; Beardsley, P.; Wilson, I.A. (2002). "Structural Insights into the Avian AICAR Transformylase Mechanism". Biochemistry. 41 (52): 15505–15513. doi:10.1021/bi020505x. PMID 12501179.
- Thoden, J.B.; Firestine, S.; Nixon, A.; Benkovic, S.J.; Holden, H.M (2000). "Molecular Structure of Escherichia coli PurT-Encoded Glycinamide Ribonucleotide Transformylase". Biochemistry. 39 (30): 8791–8802. doi:10.1021/bi000926j. PMID 10913290.
- Marolewski, A.E.; Mattia, K.M.; Warren, M.S.; Benkovic, S.J. (1997). "Formyl phosphate: a proposed intermediate in the reaction catalyzed by Escherichia coli PurT GAR transformylase". Biochemistry. 36 (22): 6709–6716. doi:10.1021/bi962961p. PMID 9184151.
- Wisniewski, J.R.; Zougman, A.; Mann, M. (2002). "N-Formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function". Nucleic Acids Research. 36 (2): 570–577. doi:10.1093/nar/gkm1057. PMC 2241850. PMID 18056081.
- Jiang, T; Zhou, X.; Taghizadeh, K.; Dong, M.; Dedon, PC. (2007). "N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage". PNAS. 104 (1): 60–65. Bibcode:2007PNAS..104...60J. doi:10.1073/pnas.0606775103. PMC 1765477. PMID 17190813.
- DeMartino, J.K.; Hwang, I.; Xu, L.; Wilson, I.A.; Boger, D.L. (2006). "Discovery of a Potent, Nonpolyglutamatable Inhibitor of Glycinamide Ribonucleotide Transformylase". Journal of Medicinal Chemistry. 49 (10): 2998–3002. doi:10.1021/jm0601147. PMC 2531195. PMID 16686541.
- Christopherson, R.I.; Lyons, S.D.; Wilson, P.K (2002). "Inhibitors of de Novo Nucleotide Biosynthesis as Drugs". Acc. Chem. Res. 35 (11): 961–971. doi:10.1021/ar0000509. PMID 12437321.
- Wang, L; Desmoulin, S.K.; Cherian, C.; Polin, L.; White, K.; Kushner, J.; Fulterer, A.; Chang, M.; Mitchell, S.; Stout, M.; Romero, M.F.; Hou, Z.; Matherly, L.H.; Gangjee, A (2011). "Synthesis, biological and antitumor activity of a highly potent 6-substituted pyrrolo[2,3-d]pyrimidine thienoyl antifolate inhibitor with proton-coupled folate transporter and folate receptor selectivity over the reduced folate carrier that inhibits β-glycinamide ribonucleotide formyltransferase". Journal of Medicinal Chemistry. 54 (20): 7150–7164. doi:10.1021/jm200739e. PMC 3209708. PMID 21879757.
- "Leigh Syndrome". Online Mendelian Inheritance in Man. Retrieved 24 February 2013.
- Tucker EJ, Hershman SG, Köhrer C, Belcher-Timme CA, Patel J, Goldberger OA, Christodoulou J, Silberstein JM, McKenzie M, Ryan MT, Compton AG, Jaffe JD, Carr SA, Calvo SE, RajBhandary UL, Thorburn DR, Mootha VK (2011). "Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation". Cell Metab. 14 (3): 428–434. doi:10.1016/j.cmet.2011.07.010. PMC 3486727. PMID 21907147.
