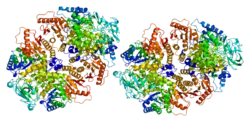
PKM2
Pyruvate kinase isozymes M1/M2 (PKM1/M2), also known as pyruvate kinase muscle isozyme (PKM), pyruvate kinase type K, cytosolic thyroid hormone-binding protein (CTHBP), thyroid hormone-binding protein 1 (THBP1), or opa-interacting protein 3 (OIP3), is an enzyme that in humans is encoded by the PKM2 gene.[5][6][7][8]
PKM2 is an isoenzyme of the glycolytic enzyme pyruvate kinase. Depending upon the different metabolic functions of the tissues, different isoenzymes of pyruvate kinase are expressed. PKM2 is expressed in some differentiated tissues, such as lung, fat tissue, retina, and pancreatic islets, as well as in all cells with a high rate of nucleic acid synthesis, such as normal proliferating cells, embryonic cells, and especially tumor cells.[9][10][11][12][13][14][15]
Structure
Two isozymes are encoded by the PKM gene: PKM1 and PKM2. The M-gene consists of 12 exons and 11 introns. PKM1 and PKM2 are different splicing products of the M-gene (exon 9 for PKM1 and exon 10 for PKM2) and solely differ in 23 amino acids within a 56-amino acid stretch (aa 378-434) at their carboxy terminus.[16][17]
Function
Pyruvate kinase catalyzes the last step within glycolysis, the dephosphorylation of phosphoenolpyruvate to pyruvate, and is responsible for net ATP production within the glycolytic sequence. In contrast to mitochondrial respiration, energy regeneration by pyruvate kinase is independent from oxygen supply and allows survival of the organs under hypoxic conditions often found in solid tumors.[18]
The involvement of this enzyme in a variety of pathways, protein–protein interactions, and nuclear transport suggests its potential to perform multiple nonglycolytic functions with diverse implications, although multidimensional role of this protein is as yet not fully explored. However, a functional role in angiogenesis the so-called process of blood vessel formation by interaction and regulation of Jmjd8 has been shown.[19][20]
Localization
Tissue
The PKM1 isozyme is expressed in organs that are strongly dependent upon a high rate of energy regeneration, such as muscle and brain.[21][22][23]
Cellular
PKM2 is enzyme pyruvate kinase M2 (PKM2) and a transcriptional coactivator of STAT1 responsible for the induction of the protein PDL-1expression and its regulation in tumor and immune cells.[24] In the lactate production, the upregulated PKM2 is required and it leads to its contribution in inflammatory response, organ injury and septic death [25][26][27] As a consequence, the removal of PKM2 in myeloid cells, administration of anti-PD-L1 or supplementation with recombinant interleukin -1 (IL-7) eases the microbial clearance, inhibits T cell apoptosis, reduce multiple organ dysfunction and reduce septic death in Bmal1-deficient mice.[28]
Subcellular
PKM2 is a cytosolic enzyme that is associated with other glycolytic enzymes, i.e., hexokinase, glyceraldehyde 3-P dehydrogenase, phosphoglycerate kinase, phosphoglyceromutase, enolase, and lactate dehydrogenase within a so-called glycolytic enzyme complex.[23][29][30][31]
However, PKM2 contains an inducible nuclear localization signal in its C-terminal domain. The role of PKM2 within the nucleus is complex, since pro-proliferative but also pro-apoptotic stimuli have been described. On the one hand, nuclear PKM2 was found to participate in the phosphorylation of histone 1 by direct phosphate transfer from PEP to histone 1. On the other hand, nuclear translocation of PKM2 induced by a somatostatin analogue, H2O2, or UV light has been linked with caspase-independent programmed cell death.[32][33][34]
Clinical significance
Bi-functional role within tumors
PKM2 is expressed in most human tumors.[11][14][15] Initially, a switch from PKM1 to PKM2 expression during tumorigenesis was discussed.[35] These conclusions, however, were the result of misinterpretation of western blots that had used PKM1-expressing mouse muscle as the sole non-cancer tissue. In clinical cancer samples, solely an up-regulation of PKM2, but no cancer specificity, could be confirmed.[36]
In contrast to the closely homologous PKM1, which always occurs in a highly active tetrameric form and which is not allosterically regulated, PKM2 may occur in a tetrameric form but also in a dimeric form. The tetrameric form of PKM2 has a high affinity to its substrate phosphoenolpyruvate (PEP), and is highly active at physiological PEP concentrations. When PKM2 is mainly in the highly active tetrameric form, which is the case in differentiated tissues and most normal proliferating cells, glucose is converted to pyruvate under the production of energy. Meanwhile, the dimeric form of PKM2 is characterized by a low affinity to its substrate PEP and is nearly inactive at physiological PEP concentrations. When PKM2 is mainly in the less active dimeric form, which is the case in tumor cells, all glycolytic intermediates above pyruvate kinase accumulate and are channelled into synthetic processes, which branch off from glycolytic intermediates such as nucleic acid-, phospholipid-, and amino acid synthesis.[21][22][23] Nucleic acids, phospholipids, and amino acids are important cell building-blocks, which are greatly needed by highly proliferating cells, such as tumor cells.
Due to the key position of pyruvate kinase within glycolysis, the tetramer:dimer ratio of PKM2 determines whether glucose carbons are converted to pyruvate and lactate under the production of energy (tetrameric form) or channelled into synthetic processes (dimeric form).[21][22][23] However, even if PKM2 activty is low leading to the diversion of upstream intermediates to synthetic processes, pyruvate and lactate will still be made using carbon atoms from glucose and other metabolites through 86 pathways bypassing pyruvate kinase.[37] These pyruvate kinase bypassing pathways are different than those participating in gluconeogenesis. Interestingly, many of the pyruvate kinase bypassing pathways use metabolites that transit through mitochondria, highlighting the importance of mitochondria in cancer metabolism irrespective of oxidative phosphorylation.
In tumor cells, PKM2 is mainly in the dimeric form and has, therefore, been termed Tumor M2-PK. The quantification of Tumor M2-PK in plasma and stool is a tool for early detection of tumors and follow-up studies during therapy. The dimerization of PKM2 in tumor cells is induced by direct interaction of PKM2 with different oncoproteins (pp60v-src, HPV-16 E7, and A-Raf).[29][30][38][39][40] The physiological function of the interaction between PKM2 and HERC1 as well as between PKM2 and PKCdelta is unknown).[41][42] Due to the essential role of PKM2 in aerobic glycolysis (The Warburg effect) which is a dominant metabolic pathway used by cancer cells.[24] Its overcome in this pathway in macrophages may lead to better outcome in experimental sepsis.[43][44][45] Thus, PKM2 is a regulator of LPS- and tumor-induced PD-L1 expression on macrophages and dendritic cells as well as tumor cells.[24]
However, the tetramer:dimer ratio of PKM2 is not stationary value. High levels of the glycolytic intermediate fructose 1,6-bisphosphate induce the re-association of the dimeric form of PKM2 to the tetrameric form. As a consequence, glucose is converted to pyruvate and lactate with the production of energy until fructose 1,6-bisphosphate levels drop below a critical value to allow dissociation to the dimeric form. This regulation is termed metabolic budget system.[22][23][46] Another activator of PKM2 is the amino acid serine.[22] The thyroid hormone 3,3´,5-triiodi-L-tyhronine (T3) binds to the monomeric form of PKM2 and prevents its association to the tetrameric form.[47]
In tumor cells, the increased rate of lactate production in the presence of oxygen is termed the Warburg effect. Genetic manipulation of cancer cells so that they produce adult PKM1 instead of PKM2 reverses the Warburg effect and reduces the growth rate of these modified cancer cells.[35] Accordingly, cotransfection of NIH 3T3 cells with gag-A-Raf and a kinase dead mutant of PKM2 reduced colony whereas cotransfection with gag-A-Raf and wild type PKM2 led to a doubling of focus formation.[48]
The dimeric form of PKM2 has been observed to have protein kinase activity in tumor cells. It is able to bind to and phosphorylate the histone H3 of chromatin in cancer cells, thereby having a role in the regulation of gene expression.[49] This modification of histone H3 and the resulting involvement in gene expression regulation can be a cause of tumor cell proliferation.[49]
The pyruvate kinase activity of PKM2 can be promoted by SAICAR (succinylaminoimidazolecarboxamide ribose-5′-phosphate), an intermediate in purine biosynthesis. In cancer cells, glucose starvation leads to a rise in SAICAR levels and the subsequent stimulation of pyruvate kinase activity of PKM2. This allows for the completion of the glycolytic pathway to produce pyruvate and, therefore, survival under glucose deprivation.[50] In addition, an abundance of SAICAR can modify glucose absorption and lactate production in cancer cells.[50] However, it has been shown that SAICAR binding also sufficiently stimulates the protein kinase activity of PKM2 in tumor cells.[51] In turn, the SAICAR-PKM2 complex can potentially phosphorylate a number of other protein kinases using PEP as the phosphate donor. Many of these proteins contribute to the regulation of cancer cell proliferation. Specifically, PKM2 can be a component in mitogen-activated protein kinase (MAPK) signaling, which is associated with increased cell proliferation if functioning improperly. This provides a potential link between SAICAR-activated PKM2 and cancer cell growth.[51]
Natural mutations and carcinogenesis
Two missense mutations, H391Y and K422R, of PKM2 were found in cells from Bloom syndrome patients prone to developing cancer. Results show that, despite the presence of mutations in the inter-subunit contact domain, the K422R and H391Y mutant proteins maintained their homotetrameric structure, similar to the wild-type protein, but showed a loss of activity of 75 and 20%, respectively. H391Y showed a 6-fold increase in affinity for its substrate phosphoenolpyruvate and behaved like a non-allosteric protein with compromised cooperative binding. However, the affinity for phosphoenolpyruvate was lost significantly in K422R. Unlike K422R, H391Y showed enhanced thermal stability, stability over a range of pH values, a lesser effect of the allosteric inhibitor Phe, and resistance toward structural alteration upon binding of the activator (fructose 1,6-bisphosphate) and inhibitor (Phe). Both mutants showed a slight shift in the pH optimum from 7.4 to 7.0.[52] The co-expression of homotetrameric wild type and mutant PKM2 in the cellular milieu resulting in the interaction between the two at the monomer level was substantiated further by in vitro experiments. The cross-monomer interaction significantly altered the oligomeric state of PKM2 by favoring dimerisation and heterotetramerization. In silico study provided an added support in showing that hetero-oligomerization was energetically favorable. The hetero-oligomeric populations of PKM2 showed altered activity and affinity, and their expression resulted in an increased growth rate of Escherichia coli as well as mammalian cells, along with an increased rate of polyploidy. These features are known to be essential to tumor progression.[53]
Further, cells stably expressing exogenous wild- or mutant-PKM2 (K422R or H391Y) or co-expressing both wild and mutant (PKM2-K422R or PKM2-H391Y), were assessed for cancer metabolism and tumorigenic potential. Cells co-expressing PKM2 and mutant (K422R or H391Y) showed significantly aggressive cancer metabolism, compared to cells expressing either wild or mutant PKM2 independently. A similar trend was observed for oxidative endurance, tumorigenic potential, cellular proliferation and tumor growth. These observations signify the dominant negative nature of these mutations. Remarkably, PKM2-H391Y co-expressed cells showed a maximal effect on all the studied parameters. Such a dominant negative impaired function of PKM2 in tumor development is not known; also evidencing for the first time the possible predisposition of BS patients with impaired PKM2 activity to cancer, and the importance of studying genetic variations in PKM2 in future to understand their relevance in cancer in general.[54]
Regulatory circuits
Cancer cells are characterized by a reprogramming of energy metabolism. Over the last decade, understanding of the metabolic changes that occur in cancer has increased dramatically, and there is great interest in targeting metabolism for cancer therapy. PKM2 plays a key role in modulating glucose metabolism to support cell proliferation. PKM2, like other PK isoforms, catalyzes the last energy-generating step in glycolysis, but is unique in its capacity to be regulated. PKM2 is regulated on several cellular levels, including gene expression, alternative splicing and post-translational modification. In addition, PKM2 is regulated by key metabolic intermediates and interacts with more than twenty different proteins. Hence, this isoenzyme is an important regulator of glycolysis and additional functions in other novel roles that have recently emerged. Recent evidence indicates that intervening in the complex regulatory network of PKM2 has severe consequences on tumor cell proliferation, indicating the potential of this enzyme as a target for tumor therapy.[55]
Bacterial pathogenesis
With the yeast two-hybrid system, gonococcal Opa proteins were found to interact with PKM2. The results suggest that direct molecular interaction with the host metabolic enzyme PKM2 is required for the acquisition of pyruvate and for gonococcal growth and survival.[56]
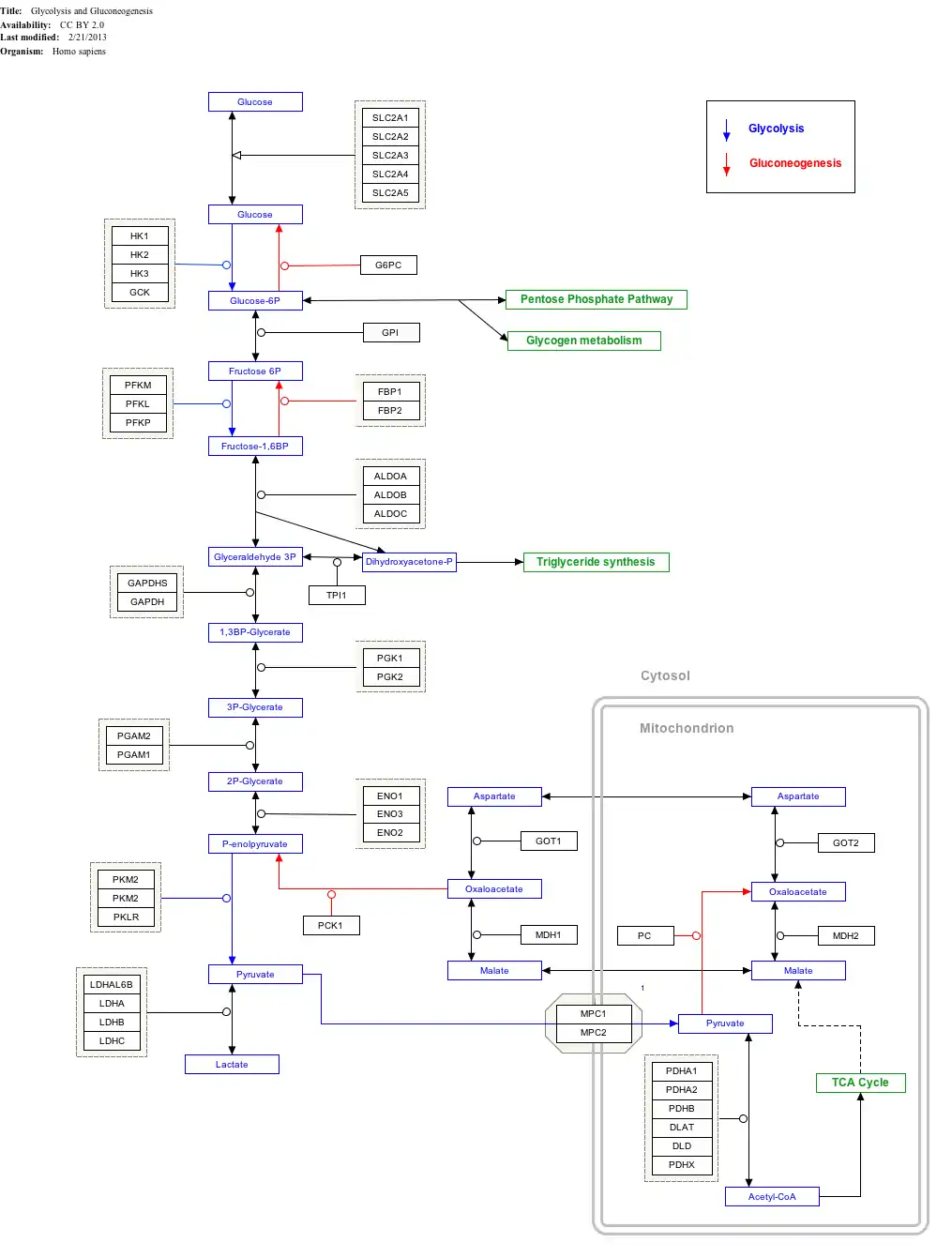
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
See also
References
- GRCh38: Ensembl release 89: ENSG00000067225 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000032294 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Kitagawa S, Obata T, Hasumura S, Pastan I, Cheng SY (March 1987). "A cellular 3,3',5-triiodo-L-thyronine binding protein from a human carcinoma cell line. Purification and characterization". The Journal of Biological Chemistry. 262 (8): 3903–8. PMID 3818670.
- Tsutsumi H, Tani K, Fujii H, Miwa S (January 1988). "Expression of L- and M-type pyruvate kinase in human tissues". Genomics. 2 (1): 86–9. doi:10.1016/0888-7543(88)90112-7. PMID 2838416.
- Tani K, Yoshida MC, Satoh H, Mitamura K, Noguchi T, Tanaka T, Fujii H, Miwa S (December 1988). "Human M2-type pyruvate kinase: cDNA cloning, chromosomal assignment and expression in hepatoma". Gene. 73 (2): 509–16. doi:10.1016/0378-1119(88)90515-X. PMID 2854097.
- Popescu NC, Cheng SY (November 1990). "Chromosomal localization of the gene for a human cytosolic thyroid hormone binding protein homologous to the subunit of pyruvate kinase, subtype M2". Somatic Cell and Molecular Genetics. 16 (6): 593–8. doi:10.1007/BF01233100. PMID 2267632. S2CID 8182554.
- Corcoran E, Phelan JJ, Fottrell PF (September 1976). "Purification and properties of pyruvate kinase from human lung". Biochimica et Biophysica Acta (BBA) - Protein Structure. 446 (1): 96–104. doi:10.1016/0005-2795(76)90101-x. PMID 974119.
- Tolle SW, Dyson RD, Newburgh RW, Cardenas JM (December 1976). "Pyruvate kinase isozymes in neurons, glia, neuroblastoma, and glioblastoma". Journal of Neurochemistry. 27 (6): 1355–1360. doi:10.1111/j.1471-4159.1976.tb02615.x. PMID 1003209.
- Reinacher M, Eigenbrodt E (1981). "Immunohistological demonstration of the same type of pyruvate kinase isoenzyme (M2-Pk) in tumors of chicken and rat". Virchows Archiv. B, Cell Pathology Including Molecular Pathology. 37 (1): 79–88. doi:10.1007/BF02892557. PMID 6116351. S2CID 34155302.
- Schering B, Eigenbrodt E, Linder D, Schoner W (August 1982). "Purification and properties of pyruvate kinase type M2 from rat lung". Biochimica et Biophysica Acta (BBA) - General Subjects. 717 (2): 337–47. doi:10.1016/0304-4165(82)90188-X. PMID 7115773.
- MacDonald MJ, Chang CM (October 1985). "Pancreatic islets contain the M2 isoenzyme of pyruvate kinase. Its phosphorylation has no effect on enzyme activity". Molecular and Cellular Biochemistry. 68 (2): 115–20. doi:10.1007/bf00219375. PMID 3908905. S2CID 6187554.
- Brinck U, Eigenbrodt E, Oehmke M, Mazurek S, Fischer G (1994). "L- and M2-pyruvate kinase expression in renal cell carcinomas and their metastases". Virchows Archiv. 424 (2): 177–85. doi:10.1007/BF00193498. PMID 8180780. S2CID 5550950.
- Steinberg P, Klingelhöffer A, Schäfer A, Wüst G, Weisse G, Oesch F, Eigenbrodt E (March 1999). "Expression of pyruvate kinase M2 in preneoplastic hepatic foci of N-nitrosomorpholine-treated rats". Virchows Archiv. 434 (3): 213–20. doi:10.1007/s004280050330. PMID 10190300. S2CID 28167108.
- Noguchi T, Inoue H, Tanaka T (October 1986). "The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing". The Journal of Biological Chemistry. 261 (29): 13807–12. PMID 3020052.
- Dombrauckas JD, Santarsiero BD, Mesecar AD (July 2005). "Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis". Biochemistry. 44 (27): 9417–29. doi:10.1021/bi0474923. PMID 15996096.
- Vaupel P, Harrison L (2004). "Tumor hypoxia: causative factors, compensatory mechanisms, and cellular response". The Oncologist. 9 Suppl 5: 4–9. doi:10.1634/theoncologist.9-90005-4. PMID 15591417.
- Gupta V, Bamezai RN (November 2010). "Human pyruvate kinase M2: a multifunctional protein". Protein Science. 19 (11): 2031–44. doi:10.1002/pro.505. PMC 3005776. PMID 20857498.
- Boeckel JN, Derlet A, Glaser SF, Luczak A, Lucas T, Heumüller AW, Krüger M, Zehendner CM, Kaluza D, Doddaballapur A, Ohtani K, Treguer K, Dimmeler S (July 2016). "JMJD8 Regulates Angiogenic Sprouting and Cellular Metabolism by Interacting With Pyruvate Kinase M2 in Endothelial Cells". Arteriosclerosis, Thrombosis, and Vascular Biology. 36 (7): 1425–33. doi:10.1161/ATVBAHA.116.307695. PMID 27199445.
- Eigenbrodt E, Glossmann H (1980). "Glycolysis – one of the keys to cancer". Trends Pharmacol. Sci. 1 (2): 240–245. doi:10.1016/0165-6147(80)90009-7.
- Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R (1992). "Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells". Critical Reviews in Oncogenesis. 3 (1–2): 91–115. PMID 1532331.
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E (August 2005). "Pyruvate kinase type M2 and its role in tumor growth and spreading". Seminars in Cancer Biology. 15 (4): 300–8. doi:10.1016/j.semcancer.2005.04.009. PMID 15908230.
- Palsson-McDermott EM, Dyck L, Zasłona Z, Menon D, McGettrick AF, Mills KH, O'Neill LA (2017-10-13). "Pyruvate Kinase M2 Is Required for the Expression of the Immune Checkpoint PD-L1 in Immune Cells and Tumors". Frontiers in Immunology. 8: 1300. doi:10.3389/fimmu.2017.01300. PMC 5646285. PMID 29081778.
- Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, Jiang JK, Jiang JK, Israelsen WJ, Keane J, Thomas C, Clish C, Vander Heiden M, Vanden Heiden M, Xavier RJ, O'Neill LA (January 2015). "Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages". Cell Metabolism. 21 (1): 65–80. doi:10.1016/j.cmet.2014.12.005. PMC 5198835. PMID 25565206.
- Zhang Z, Deng W, Kang R, Xie M, Billiar T, Wang H, Cao L, Tang D (September 2016). "Plumbagin Protects Mice from Lethal Sepsis by Modulating Immunometabolism Upstream of PKM2". Molecular Medicine. 22: 162–172. doi:10.2119/molmed.2015.00250. PMC 5004715. PMID 26982513.
- Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, Kang R, Lotze MT, Billiar TR, Wang H, Cao L, Tang D (July 2014). "PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis". Nature Communications. 5 (1): 4436. Bibcode:2014NatCo...5.4436Y. doi:10.1038/ncomms5436. PMC 4104986. PMID 25019241.
- Deng W, Zhu S, Zeng L, Liu J, Kang R, Yang M, Cao L, Wang H, Billiar TR, Jiang J, Xie M, Tang D (July 2018). "The Circadian Clock Controls Immune Checkpoint Pathway in Sepsis". Cell Reports. 24 (2): 366–378. doi:10.1016/j.celrep.2018.06.026. PMC 6094382. PMID 29996098.
- Zwerschke W, Mazurek S, Massimi P, Banks L, Eigenbrodt E, Jansen-Dürr P (February 1999). "Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein". Proceedings of the National Academy of Sciences of the United States of America. 96 (4): 1291–6. Bibcode:1999PNAS...96.1291Z. doi:10.1073/pnas.96.4.1291. PMC 15456. PMID 9990017.
- Mazurek S, Zwerschke W, Jansen-Dürr P, Eigenbrodt E (October 2001). "Metabolic cooperation between different oncogenes during cell transformation: interaction between activated ras and HPV-16 E7". Oncogene. 20 (47): 6891–8. doi:10.1038/sj.onc.1204792. PMID 11687968.
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC (March 2008). "Pyruvate kinase M2 is a phosphotyrosine-binding protein". Nature. 452 (7184): 181–6. Bibcode:2008Natur.452..181C. doi:10.1038/nature06667. PMID 18337815. S2CID 4346405.
- Ignacak J, Stachurska MB (March 2003). "The dual activity of pyruvate kinase type M2 from chromatin extracts of neoplastic cells". Comparative Biochemistry and Physiology. Part B, Biochemistry & Molecular Biology. 134 (3): 425–33. doi:10.1016/S1096-4959(02)00283-X. PMID 12628374.
- Hoshino A, Hirst JA, Fujii H (June 2007). "Regulation of cell proliferation by interleukin-3-induced nuclear translocation of pyruvate kinase". The Journal of Biological Chemistry. 282 (24): 17706–11. doi:10.1074/jbc.M700094200. PMID 17446165.
- Steták A, Veress R, Ovádi J, Csermely P, Kéri G, Ullrich A (February 2007). "Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death". Cancer Research. 67 (4): 1602–8. doi:10.1158/0008-5472.CAN-06-2870. PMID 17308100.
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC (March 2008). "The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth". Nature. 452 (7184): 230–3. Bibcode:2008Natur.452..230C. doi:10.1038/nature06734. PMID 18337823. S2CID 16111842.
- Bluemlein K, Grüning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M (May 2011). "No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis". Oncotarget. 2 (5): 393–400. doi:10.18632/oncotarget.278. PMC 3248187. PMID 21789790.
- Christos Chinopoulos (2020), From Glucose to Lactate and Transiting Intermediates Through Mitochondria, Bypassing Pyruvate Kinase: Considerations for Cells Exhibiting Dimeric PKM2 or Otherwise Inhibited Kinase Activity, https://www.frontiersin.org/articles/10.3389/fphys.2020.543564/full
- Oude Weernink PA, Rijksen G, Staal GE (1991). "Phosphorylation of pyruvate kinase and glycolytic metabolism in three human glioma cell lines". Tumour Biology. 12 (6): 339–52. doi:10.1159/000217735. PMID 1798909.
- Eigenbrodt E, Mazurek S, Friis RR (1998). Double role of pyruvate kinase type M2 in the regulation of phosphometabolite pools. In: Bannasch P, Kanduc D, Papa S, Tager JM (eds). Cell Growth and Oncogenesis. Basel/Switzerland: Birkhäuser Verlag. pp. 15–30. doi:10.1007/978-3-0348-8950-6_2. ISBN 3-7643-5727-4.
- Mazurek S, Drexler HC, Troppmair J, Eigenbrodt E, Rapp UR (2007). "Regulation of pyruvate kinase type M2 by A-Raf: a possible glycolytic stop or go mechanism". Anticancer Research. 27 (6B): 3963–71. PMID 18225557.
- Garcia-Gonzalo FR, Cruz C, Muñoz P, Mazurek S, Eigenbrodt E, Ventura F, Bartrons R, Rosa JL (March 2003). "Interaction between HERC1 and M2-type pyruvate kinase". FEBS Letters. 539 (1–3): 78–84. doi:10.1016/S0014-5793(03)00205-9. PMID 12650930. S2CID 32809019.
- Siwko S, Mochly-Rosen D (2007). "Use of a novel method to find substrates of protein kinase C delta identifies M2 pyruvate kinase". The International Journal of Biochemistry & Cell Biology. 39 (5): 978–87. doi:10.1016/j.biocel.2007.01.018. PMC 1931518. PMID 17337233.
- Zhang Z, Deng W, Kang R, Xie M, Billiar T, Wang H, et al. (September 2016). "Plumbagin Protects Mice from Lethal Sepsis by Modulating Immunometabolism Upstream of PKM2". Molecular Medicine. 22: 162–172. doi:10.2119/molmed.2015.00250. PMC 5004715. PMID 26982513.
- Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, et al. (July 2014). "PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis". Nature Communications. 5: 4436. Bibcode:2014NatCo...5.4436Y. doi:10.1038/ncomms5436. PMC 4104986. PMID 25019241.
- Huang J, Liu K, Zhu S, Xie M, Kang R, Cao L, Tang D (August 2018). "AMPK regulates immunometabolism in sepsis". Brain, Behavior, and Immunity. 72: 89–100. doi:10.1016/j.bbi.2017.11.003. PMID 29109024. S2CID 38415440.
- Ashizawa K, Willingham MC, Liang CM, Cheng SY (September 1991). "In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate". The Journal of Biological Chemistry. 266 (25): 16842–6. PMID 1885610.
- Kato H, Fukuda T, Parkison C, McPhie P, Cheng SY (October 1989). "Cytosolic thyroid hormone-binding protein is a monomer of pyruvate kinase". Proceedings of the National Academy of Sciences of the United States of America. 86 (20): 7861–5. Bibcode:1989PNAS...86.7861K. doi:10.1073/pnas.86.20.7861. PMC 298171. PMID 2813362.
- Le Mellay V, Houben R, Troppmair J, Hagemann C, Mazurek S, Frey U, Beigel J, Weber C, Benz R, Eigenbrodt E, Rapp UR (2002). "Regulation of glycolysis by Raf protein serine/threonine kinases". Advances in Enzyme Regulation. 42: 317–32. doi:10.1016/S0065-2571(01)00036-X. PMID 12123723.
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z (August 2012). "PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis". Cell. 150 (4): 685–96. doi:10.1016/j.cell.2012.07.018. PMC 3431020. PMID 22901803.
- Keller KE, Tan IS, Lee YS (November 2012). "SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions". Science. 338 (6110): 1069–72. Bibcode:2012Sci...338.1069K. doi:10.1126/science.1224409. PMC 3527123. PMID 23086999.
- Keller KE, Doctor ZM, Dwyer ZW, Lee YS (March 2014). "SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells". Molecular Cell. 53 (5): 700–9. doi:10.1016/j.molcel.2014.02.015. PMC 4000728. PMID 24606918.
- Akhtar K, Gupta V, Koul A, Alam N, Bhat R, Bamezai RN (May 2009). "Differential behavior of missense mutations in the intersubunit contact domain of the human pyruvate kinase M2 isozyme". The Journal of Biological Chemistry. 284 (18): 11971–81. doi:10.1074/jbc.M808761200. PMC 2673266. PMID 19265196.
- Gupta V, Kalaiarasan P, Faheem M, Singh N, Iqbal MA, Bamezai RN (May 2010). "Dominant negative mutations affect oligomerization of human pyruvate kinase M2 isozyme and promote cellular growth and polyploidy". The Journal of Biological Chemistry. 285 (22): 16864–73. doi:10.1074/jbc.M109.065029. PMC 2878009. PMID 20304929.
- Iqbal MA, Siddiqui FA, Chaman N, Gupta V, Kumar B, Gopinath P, Bamezai RN (March 2014). "Missense mutations in pyruvate kinase M2 promote cancer metabolism, oxidative endurance, anchorage independence, and tumor growth in a dominant negative manner". The Journal of Biological Chemistry. 289 (12): 8098–105. doi:10.1074/jbc.M113.515742. PMC 3961641. PMID 24492614.
- Gupta V, Wellen KE, Mazurek S, Bamezai RN (2013). "Pyruvate kinase M2: regulatory circuits and potential for therapeutic intervention". Current Pharmaceutical Design. 20 (15): 2595–606. doi:10.2174/13816128113199990484. PMID 23859618.
- Williams JM, Chen GC, Zhu L, Rest RF (January 1998). "Using the yeast two-hybrid system to identify human epithelial cell proteins that bind gonococcal Opa proteins: intracellular gonococci bind pyruvate kinase via their Opa proteins and require host pyruvate for growth". Molecular Microbiology. 27 (1): 171–86. doi:10.1046/j.1365-2958.1998.00670.x. PMID 9466265.
External links
- Pyruvate+kinase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Erich Eigenbrodt; Sybille Mazurek. "Pyruvate kinase isoenzyme type M2 (M2-PK)". Tumor metabolome database. Retrieved 2008-03-22.
- Overview of all the structural information available in the PDB for UniProt: P14618 (Pyruvate kinase PKM) at the PDBe-KB.