Lactate dehydrogenase
Lactate dehydrogenase (LDH or LD) is an enzyme found in nearly all living cells. LDH catalyzes the conversion of lactate to pyruvate and back, as it converts NAD+ to NADH and back. A dehydrogenase is an enzyme that transfers a hydride from one molecule to another.
| Lactate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Lactate dehydrogenase M tetramer (LDH5), Human | |||||||||
| Identifiers | |||||||||
| EC number | 1.1.1.27 | ||||||||
| CAS number | 9001-60-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
LDH exists in four distinct enzyme classes. This article is specifically about the NAD(P)-dependent L-lactate dehydrogenase. Other LDHs act on D-lactate and/or are dependent on cytochrome c: D-lactate dehydrogenase (cytochrome) and L-lactate dehydrogenase (cytochrome).
LDH is expressed extensively in body tissues, such as blood cells and heart muscle. Because it is released during tissue damage, it is a marker of common injuries and disease such as heart failure.
Reaction

Lactate dehydrogenase catalyzes the interconversion of pyruvate and lactate with concomitant interconversion of NADH and NAD+. It converts pyruvate, the final product of glycolysis, to lactate when oxygen is absent or in short supply, and it performs the reverse reaction during the Cori cycle in the liver. At high concentrations of lactate, the enzyme exhibits feedback inhibition, and the rate of conversion of pyruvate to lactate is decreased. It also catalyzes the dehydrogenation of 2-hydroxybutyrate, but this is a much poorer substrate than lactate.
Active site
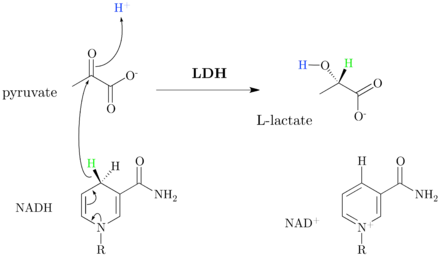
LDH in humans uses His(193) as the proton donor, and works in unison with the coenzyme (Arg99 and Asn138), and substrate (Arg106; Arg169; Thr248) binding residues.[1][2] The His(193) active site, is not only found in the human form of LDH, but is found in many different animals, showing the convergent evolution of LDH. The two different subunits of LDH (LDHA, also known as the M subunit of LDH, and LDHB, also known as the H subunit of LDH) both retain the same active site and the same amino acids participating in the reaction. The noticeable difference between the two subunits that make up LDH's tertiary structure is the replacement of alanine (in the M chain) with a glutamine (in the H chain). This tiny but notable change is believed to be the reason the H subunit can bind faster, and the M subunit's catalytic activity isn't reduced when subjected to the same conditions as the H subunit, whereas the H subunit's activity is reduced fivefold.[3]
Isozymes
Lactate dehydrogenase is composed of four subunits (tetramer). The two most common subunits are the LDH-M and LDH-H protein, encoded by the LDHA and LDHB genes, respectively. These two subunits can form five possible tetramers (isoenzymes): 4H, 4M, and the three mixed tetramers (3H1M, 2H2M, 1H3M). These five isoforms are enzymatically similar but show different tissue distribution: The major isoenzymes of skeletal muscle and liver, M4, has four muscle (M) subunits, while H4 is the main isoenzymes for heart muscle in most species, containing four heart (H) subunits.
- LDH-1 (4H)—in the heart and in RBC (red blood cells), as well as the brain[4]
- LDH-2 (3H1M)—in the reticuloendothelial system
- LDH-3 (2H2M)—in the lungs
- LDH-4 (1H3M)—in the kidneys, placenta, and pancreas
- LDH-5 (4M)—in the liver and striated muscle,[5] also present in the brain[4]
LDH-2 is usually the predominant form in the serum. An LDH-1 level higher than the LDH-2 level (a "flipped pattern") suggests myocardial infarction (damage to heart tissues releases heart LDH, which is rich in LDH-1, into the bloodstream). The use of this phenomenon to diagnose infarction has been largely superseded by the use of Troponin I or T measurement.
There are two more mammalian LDH subunits that can be included in LDH tetramers: LDHC and LDHBx. LDHC is a testes-specific LDH protein, that is encoded by the LDHC gene. LDHBx is a peroxisome-specific LDH protein. LDHBx is the readthrough-form of LDHB. LDHBx is generated by translation of the LDHB mRNA, but the stop codon is interpreted as an amino acid-encoding codon. In consequence, translation continues to the next stop codon. This leads to the addition of seven amino acid residues to the normal LDH-H protein. The extension contains a peroxisomal targeting signal, so that LDHBx is imported into the peroxisome.[6]
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein families
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The family also contains L-lactate dehydrogenases that catalyse the conversion of L-lactate to pyruvate, the last step in anaerobic glycolysis. Malate dehydrogenases that catalyse the interconversion of malate to oxaloacetate and participate in the citric acid cycle, and L-2-hydroxyisocaproate dehydrogenases are also members of the family. The N-terminus is a Rossmann NAD-binding fold and the C-terminus is an unusual alpha+beta fold.[7][8]
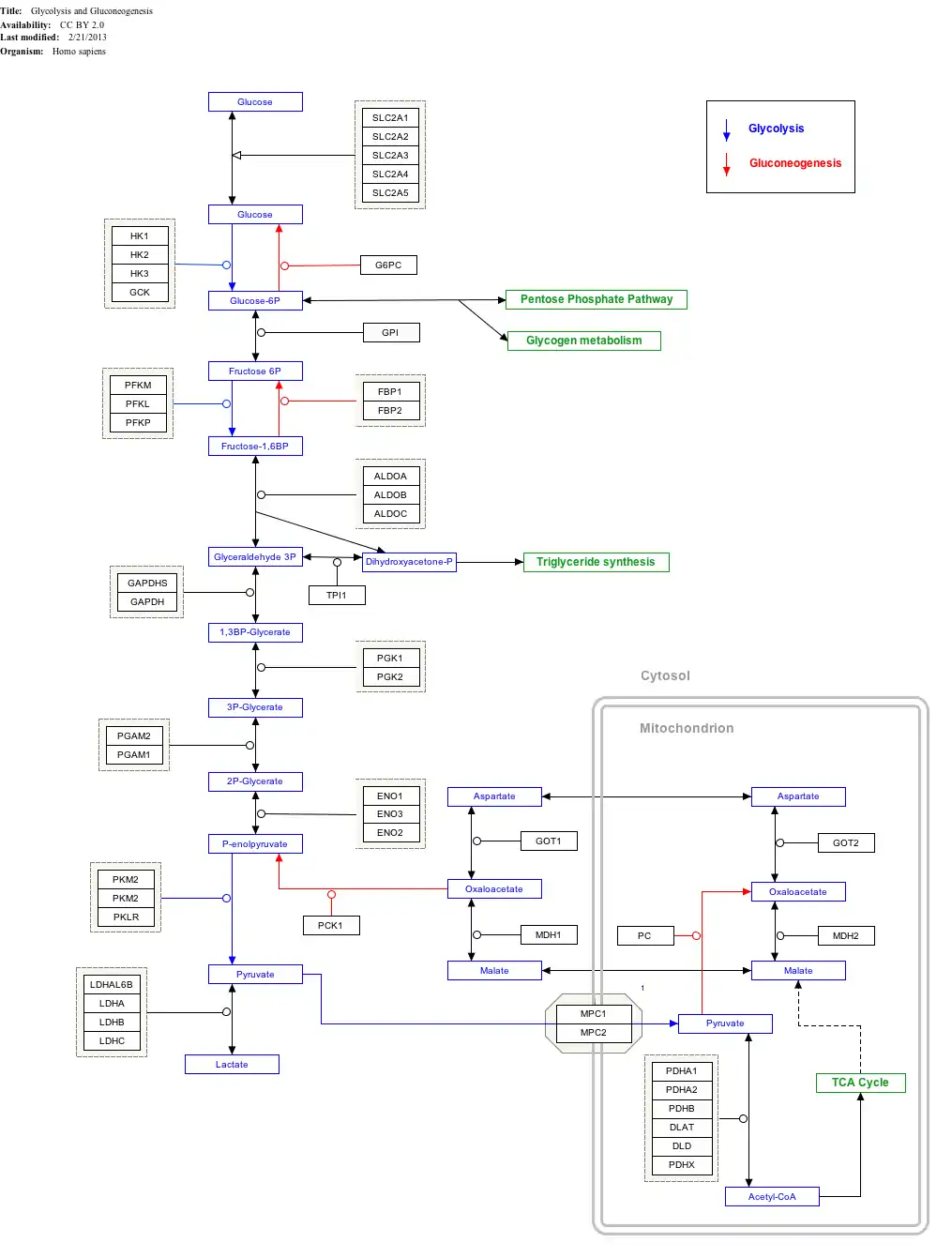
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
Enzyme regulation
This protein may use the morpheein model of allosteric regulation.[9]
Ethanol-induced hypoglycemia
Ethanol is dehydrogenated to acetaldehyde by alcohol dehydrogenase, and further into acetic acid by acetaldehyde dehydrogenase. During this reaction 2 NADH are produced. If large amounts of ethanol are present, then large amounts of NADH are produced, leading to a depletion of NAD+. Thus, the conversion of pyruvate to lactate is increased due to the associated regeneration of NAD+. Therefore, anion-gap metabolic acidosis (lactic acidosis) may ensue in ethanol poisoning.
The increased NADH/NAD+ ratio also can cause hypoglycemia in an (otherwise) fasting individual who has been drinking and is dependent on gluconeogenesis to maintain blood glucose levels. Alanine and lactate are major gluconeogenic precursors that enter gluconeogenesis as pyruvate. The high NADH/NAD+ ratio shifts the lactate dehydrogenase equilibrium to lactate, so that less pyruvate can be formed and, therefore, gluconeogenesis is impaired.
Substrate regulation
LDH is also regulated by the relative concentrations of its substrates. LDH becomes more active under periods of extreme muscular output due to an increase in substrates for the LDH reaction. When skeletal muscles are pushed to produce high levels of power, the demand for ATP in regards to aerobic ATP supply leads to an accumulation of free ADP, AMP, and Pi. The subsequent glycolytic flux, specifically production of pyruvate, exceeds the capacity for pyruvate dehydrogenase and other shuttle enzymes to metabolize pyruvate. The flux through LDH increases in response to increased levels of pyruvate and NADH to metabolize pyruvate into lactate.[10]
Transcriptional regulation
LDH undergoes transcriptional regulation by PGC-1α. PGC-1α regulates LDH by decreasing LDH A mRNA transcription and the enzymatic activity of pyruvate to lactate conversion.[11]
Genetics
The M and H subunits are encoded by two different genes:
- The M subunit is encoded by LDHA, located on chromosome 11p15.4 (Online Mendelian Inheritance in Man (OMIM): 150000).
- The H subunit is encoded by LDHB, located on chromosome 12p12.2-p12.1 (Online Mendelian Inheritance in Man (OMIM): 150100).
- A third isoform, LDHC or LDHX, is expressed only in the testis (Online Mendelian Inheritance in Man (OMIM): 150150); its gene is likely a duplicate of LDHA and is also located on the eleventh chromosome (11p15.5-p15.3).
- The fourth isoform is localized in the peroxisome. It is tetramer containing one LDHBx subunit, which is also encoded by the LDHB gene. The LDHBx protein is seven amino acids longer than the LDHB (LDH-H) protein. This amino acid extension is generated by functional translational readthrough.[6]
Mutations of the M subunit have been linked to the rare disease exertional myoglobinuria (see OMIM article), and mutations of the H subunit have been described but do not appear to lead to disease.
Mutations
In rare cases, a mutation in the genes controlling the production of lactate dehydrogenase will lead to a medical condition known as lactate dehydrogenase deficiency. Depending on which gene carries the mutation, one of two types will occur: either lactate dehydrogenase-A deficiency (also known as glycogen storage disease XI) or lactate dehydrogenase-B deficiency. Both of these conditions affect how the body breaks down sugars, primarily in certain muscle cells. Lactate dehydrogenase-A deficiency is caused by a mutation to the LDHA gene, while lactate dehydrogenase-B deficiency is caused by a mutation to the LDHB gene.[12]
This condition is inherited in an autosomal recessive pattern, meaning that both parents must contribute a mutated gene in order for this condition to be expressed.[13]
A complete lactate dehydrogenase enzyme consists of four protein subunits.[14] Since the two most common subunits found in lactate dehydrogenase are encoded by the LDHA and LDHB genes, either variation of this disease causes abnormalities in many of the lactate dehydrogenase enzymes found in the body. In the case of lactate dehydrogenase-A deficiency, mutations to the LDHA gene result in the production of an abnormal lactate dehydrogenase-A subunit that cannot bind to the other subunits to form the complete enzyme. This lack of a functional subunit reduces the amount of enzyme formed, leading to an overall decrease in activity. During the anaerobic phase of glycolysis (the Cori Cycle), the mutated enzyme is unable to convert pyruvate into lactate to produce the extra energy the cells need. Since this subunit has the highest concentration in the LDH enzymes found in the skeletal muscles (which are the primary muscles responsible for movement), high-intensity physical activity will lead to an insufficient amount of energy being produced during this anaerobic phase.[15] This in turn will cause the muscle tissue to weaken and eventually break down, a condition known as rhabdomyolysis. The process of rhabdomyolysis also releases myoglobin into the blood, which will eventually end up in the urine and cause it to become red or brown: another condition known as myoglobinuria.[16] Some other common symptoms are exercise intolerance, which consists of fatigue, muscle pain, and cramps during exercise, and skin rashes.[17][18] In severe cases, myoglobinuria can damage the kidneys and lead to life-threatening kidney failure.[19] In order to obtain a definitive diagnosis, a muscle biopsy may be performed to confirm low or absent LDH activity. There is currently no specific treatment for this condition.
In the case of lactate dehydrogenase-B deficiency, mutations to the LDHB gene result in the production of an abnormal lactate dehydrogenase-B subunit that cannot bind to the other subunits to form the complete enzyme. As with lactate dehydrogenase-A deficiency, this mutation reduces the overall effectiveness in the enzyme.[20] However, there are some major differences between these two cases. The first is the location where the condition manifests itself. With lactate dehydrogenase-B deficiency, the highest concentration of B subunits can be found within the cardiac muscle, or the heart. Within the heart, lactate dehydrogenase plays the role of converting lactate back into pyruvate so that the pyruvate can be used again to create more energy.[21] With the mutated enzyme, the overall rate of this conversion is decreased. However, unlike lactate dehydrogenase-A deficiency, this mutation does not appear to cause any symptoms or health problems linked to this condition.[18][22] At the present moment, it is unclear why this is the case. Affected individuals are usually discovered only when routine blood tests indicate low LDH levels present within the blood.
Role in muscular fatigue
The onset of acidosis during periods of intense exercise is commonly attributed to accumulation of hydrogens that are dissociated from lactate. Previously, lactic acid was thought to cause fatigue. From this reasoning, the idea of lactate production being a primary cause of muscle fatigue during exercise was widely adopted. A closer, mechanistic analysis of lactate production under “anaerobic” conditions shows that there is no biochemical evidence for the production of lactate through LDH contributing to acidosis. While LDH activity is correlated to muscle fatigue,[23] the production of lactate by means of the LDH complex works as a system to delay the onset of muscle fatigue. George Brooks and Colleagues at UC Berkeley where the lactate shuttle was discovered showed that lactate was actually a metabolic fuel not a waste product or the cause of fatigue.
LDH works to prevent muscular failure and fatigue in multiple ways. The lactate-forming reaction generates cytosolic NAD+, which feeds into the glyceraldehyde 3-phosphate dehydrogenase reaction to help maintain cytosolic redox potential and promote substrate flux through the second phase of glycolysis to promote ATP generation. This, in effect, provides more energy to contracting muscles under heavy workloads. The production and removal of lactate from the cell also ejects a proton consumed in the LDH reaction- the removal of excess protons produced in the wake of this fermentation reaction serves to act as a buffer system for muscle acidosis. Once proton accumulation exceeds the rate of uptake in lactate production and removal through the LDH symport,[24] muscular acidosis occurs.
Blood test
| Lower limit | Upper limit | Unit | Comments |
|---|---|---|---|
| 50[25] | 150[25] | U/L | |
| 0.4[26] | 1.7[26] | μmol/L | |
| 1.8[27] | 3.4[27] | µkat/L | < 70 years old[27] |
On blood tests, an elevated level of lactate dehydrogenase usually indicates tissue damage, which has multiple potential causes, reflecting its widespread tissue distribution:
- Hemolytic anemia[28]
- Vitamin B12 deficiency anemia[28]
- Infections such as infectious mononucleosis, meningitis, encephalitis, HIV/AIDS. It is notably increased in sepsis.[28]
- Infarction, such as bowel infarction, myocardial infarction and lung infarction[28]
- Acute kidney disease[28]
- Acute liver disease[28]
- Rhabdomyolysis[29]
- Pancreatitis[28]
- Bone fractures[28]
- Cancers, notably testicular cancer and lymphoma. A high LDH after chemotherapy indicates a poorer response and a worse survival rate.[28]
- Severe shock[28]
- Hypoxia[28]
Low and normal levels of LDH do not usually indicate any pathology.[28] Low levels may be caused by large intake of vitamin C.
LDH is a protein that normally appears throughout the body in small amounts.
Testing in cancer
Many cancers can raise LDH levels, so LDH may be used as a tumor marker, but at the same time, it is not useful in identifying a specific kind of cancer. Measuring LDH levels can be helpful in monitoring treatment for cancer. Noncancerous conditions that can raise LDH levels include heart failure, hypothyroidism, anemia, pre-eclampsia, meningitis, encephalitis, acute pancreatitis, HIV and lung or liver disease.[30]
Tissue breakdown releases LDH, and therefore, LDH can be measured as a surrogate for tissue breakdown (e.g., hemolysis). LDH is measured by the lactate dehydrogenase (LDH) test (also known as the LDH test or lactic acid dehydrogenase test). Comparison of the measured LDH values with the normal range help guide diagnosis.[31]
Hemolysis
In medicine, LDH is often used as a marker of tissue breakdown as LDH is abundant in red blood cells and can function as a marker for hemolysis. A blood sample that has been handled incorrectly can show false-positively high levels of LDH due to erythrocyte damage.
It can also be used as a marker of myocardial infarction. Following a myocardial infarction, levels of LDH peak at 3–4 days and remain elevated for up to 10 days. In this way, elevated levels of LDH (where the level of LDH1 is higher than that of LDH2, i.e. the LDH Flip, as normally, in serum, LDH2 is higher than LDH1) can be useful for determining whether a patient has had a myocardial infarction if they come to doctors several days after an episode of chest pain.
Tissue turnover
Other uses are assessment of tissue breakdown in general; this is possible when there are no other indicators of hemolysis. It is used to follow up cancer (especially lymphoma) patients, as cancer cells have a high rate of turnover, with destroyed cells leading to an elevated LDH activity.
HIV
LDH is often measured in HIV patients as a non-specific marker for pneumonia due to Pneumocystis jirovecii (PCP). Elevated LDH in the setting of upper respiratory symptoms in a HIV patient suggests, but is not diagnostic for, PCP. However, in HIV-positive patients with respiratory symptoms, a very high LDH level (>600 IU/L) indicated histoplasmosis (9.33 times more likely) in a study of 120 PCP and 30 histoplasmosis patients.[32]
Testing in other body fluids
Exudates and transudates
Measuring LDH in fluid aspirated from a pleural effusion (or pericardial effusion) can help in the distinction between exudates (actively secreted fluid, e.g., due to inflammation) or transudates (passively secreted fluid, due to a high hydrostatic pressure or a low oncotic pressure). The usual criterion (included in Light's criteria) is that a ratio of fluid LDH versus upper limit of normal serum LDH of more than 0.6[33] or 2⁄3[34] indicates an exudate, while a ratio of less indicates a transudate. Different laboratories have different values for the upper limit of serum LDH, but examples include 200[35] and 300[35] IU/L.[36] In empyema, the LDH levels, in general, will exceed 1000 IU/L.
Meningitis and encephalitis
High levels of lactate dehydrogenase in cerebrospinal fluid are often associated with bacterial meningitis.[37] In the case of viral meningitis, high LDH, in general, indicates the presence of encephalitis and poor prognosis.
In cancer treatment
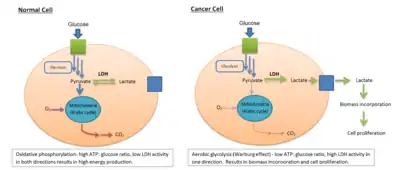
LDH is involved in tumor initiation and metabolism. Cancer cells rely on increased glycolysis resulting in increased lactate production in addition to aerobic respiration in the mitochondria, even under oxygen-sufficient conditions (a process known as the Warburg effect[38]). This state of fermentative glycolysis is catalyzed by the A form of LDH. This mechanism allows tumorous cells to convert the majority of their glucose stores into lactate regardless of oxygen availability, shifting use of glucose metabolites from simple energy production to the promotion of accelerated cell growth and replication.
LDH A and the possibility of inhibiting its activity has been identified as a promising target in cancer treatments focused on preventing carcinogenic cells from proliferating. Chemical inhibition of LDH A has demonstrated marked changes in metabolic processes and overall survival of carcinoma cells. Oxamate is a cytosolic inhibitor of LDH A that significantly decreases ATP production in tumorous cells as well as increasing production of reactive oxygen species (ROS). These ROS drive cancer cell proliferation by activating kinases that drive cell cycle progression growth factors at low concentrations,[39] but can damage DNA through oxidative stress at higher concentrations. Secondary lipid oxidation products can also inactivate LDH and impact its ability to regenerate NADH,[40] directly disrupting the enzymes ability to convert lactate to pyruvate.
While recent studies have shown that LDH activity is not necessarily an indicator of metastatic risk,[41] LDH expression can act as a general marker in the prognosis of cancers. Expression of LDH5 and VEGF in tumors and the stroma has been found to be a strong prognostic factor for diffuse or mixed-type gastric cancers.[42]
Prokaryotes
A cap-membrane-binding domain is found in prokaryotic lactate dehydrogenase. This consists of a large seven-stranded antiparallel beta-sheet flanked on both sides by alpha-helices. It allows for membrane association.[43]
See also
References
- Holmes RS, Goldberg E (October 2009). "Computational analyses of mammalian lactate dehydrogenases: human, mouse, opossum and platypus LDHs". Computational Biology and Chemistry. 33 (5): 379–85. doi:10.1016/j.compbiolchem.2009.07.006. PMC 2777655. PMID 19679512.
- Wilks HM, Holbrook JJ (December 1988). "A Specific, Highly Active Malate Dehydrogenase by Redesign of a Lactate Dehydrogenase Framework". Science. 242 (4885): 1541–44. Bibcode:1988Sci...242.1541W. doi:10.1126/science.3201242. PMID 3201242.
- Eventoff W, Rossmann MG, Taylor SS, Torff HJ, Meyer H, Keil W, Kiltz HH (July 1977). "Structural adaptations of lactate dehydrogenase isozymes". Proceedings of the National Academy of Sciences of the United States of America. 74 (7): 2677–81. Bibcode:1977PNAS...74.2677E. doi:10.1073/pnas.74.7.2677. PMC 431242. PMID 197516.
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. (September 2014). "An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex". The Journal of Neuroscience. 34 (36): 11929–47. doi:10.1523/JNEUROSCI.1860-14.2014. PMC 4152602. PMID 25186741.
- Van Eerd JP, Kreutzer EK (1996). Klinische Chemie voor Analisten deel 2. pp. 138–139. ISBN 978-90-313-2003-5.
- Schueren F, Lingner T, George R, Hofhuis J, Gartner J, Thoms S (2014). "Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals". eLife. 3: e03640. doi:10.7554/eLife.03640. PMC 4359377. PMID 25247702.
- Chapman AD, Cortés A, Dafforn TR, Clarke AR, Brady RL (1999). "Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD". J Mol Biol. 285 (2): 703–12. doi:10.1006/jmbi.1998.2357. PMID 10075524.
- Madern D (2002). "Molecular evolution within the L-malate and L-lactate dehydrogenase super-family". J Mol Evol. 54 (6): 825–40. Bibcode:2002JMolE..54..825M. doi:10.1007/s00239-001-0088-8. PMID 12029364. S2CID 469660.
- Selwood T, Jaffe EK (March 2012). "Dynamic dissociating homo-oligomers and the control of protein function". Arch. Biochem. Biophys. 519 (2): 131–43. doi:10.1016/j.abb.2011.11.020. PMC 3298769. PMID 22182754.
- Spriet LL, Howlett RA, Heigenhauser GJ (2000). "An enzymatic approach to lactate production in human skeletal muscle during exercise". Med Sci Sports Exerc. 32 (4): 756–63. doi:10.1097/00005768-200004000-00007. PMID 10776894.
- Summermatter S, Santos G, Pérez-Schindler J, Handschin C (2013). "Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A." Proc Natl Acad Sci U S A. 110 (21): 8738–43. Bibcode:2013PNAS..110.8738S. doi:10.1073/pnas.1212976110. PMC 3666691. PMID 23650363.
- "Lactate dehydrogenase deficiency". Genetics Home Reference. 29 February 2016. Retrieved 2 March 2016.
- "Diseases – Metabolic Diseases – Causes/Inheritance". Muscular Dystrophy Association. Retrieved 2 March 2016.
- Millar DB, Frattali V, Willick GE (June 1969). "The quaternary structure of lactate dehydrogenase. I. The subunit molecular weight and the reversible association at acid pH". Biochemistry. 8 (6): 2416–21. doi:10.1021/bi00834a025. PMID 5816379.
- Kanno T, Sudo K, Maekawa M, Nishimura Y, Ukita M, Fukutake K (March 1988). "Lactate dehydrogenase M-subunit deficiency: a new type of hereditary exertional myopathy". Clinica Chimica Acta; International Journal of Clinical Chemistry. 173 (1): 89–98. doi:10.1016/0009-8981(88)90359-2. PMID 3383424.
- "Myoglobinuria; Rhabdomyolysis". neuromuscular.wustl.edu. Retrieved 2 March 2016.
- Hoffmann GF (1 January 2002). Inherited Metabolic Diseases. Lippincott Williams & Wilkins. ISBN 9780781729000.
- "Glycogenoses". neuromuscular.wustl.edu. Retrieved 2 March 2016.
- "Glycogen storage disease XI – Conditions – GTR – NCBI". www.ncbi.nlm.nih.gov. Retrieved 2 March 2016.
- "LDHB gene". Genetics Home Reference. 29 February 2016. Retrieved 2 March 2016.
- "Lactate Dehydrogenase – Worthington Enzyme Manual". www.worthington-biochem.com. Retrieved 2 March 2016.
- "OMIM Entry # 614128 – LACTATE DEHYDROGENASE B DEFICIENCY; LDHBD". www.omim.org. Retrieved 2 March 2016.
- Tesch P, Sjödin B, Thorstensson A, Karlsson J (1978). "Muscle fatigue and its relation to lactate accumulation and LDH activity in man". Acta Physiol Scand. 103 (4): 413–20. doi:10.1111/j.1748-1716.1978.tb06235.x. PMID 716962.
- Juel C, Klarskov C, Nielsen JJ, Krustrup P, Mohr M, Bangsbo J (2004). "Effect of high-intensity intermittent training on lactate and H+ release from human skeletal muscle". Am J Physiol Endocrinol Metab. 286 (2): E245-51. doi:10.1152/ajpendo.00303.2003. PMID 14559724.
- Blood Test Results – Normal Ranges Archived 2012-11-02 at the Wayback Machine Bloodbook.Com
- Fachwörterbuch Kompakt Medizin E-D/D-E. Author: Fritz-Jürgen Nöhring. Edition 2. Publisher:Elsevier, Urban&FischerVerlag, 2004. ISBN 3-437-15120-7, ISBN 978-3-437-15120-0. Length: 1288 pages
- Reference range list from Uppsala University Hospital ("Laborationslista"). Artnr 40284 Sj74a. Issued on April 22, 2008
- "Lactate Dehydrogenase (LD)". Lab Tests Online. Last reviewed on June 22, 2018.}}
- Keltz E, Khan FY, Mann G (2019). "Rhabdomyolysis. The role of diagnostic and prognostic factors". Muscles Ligaments Tendons J. 3 (4): 303–12. doi:10.32098/mltj.04.2013.11. PMC 3940504. PMID 24596694.CS1 maint: multiple names: authors list (link)
- Stanford Cancer Center. "Cancer Diagnosis – Understanding Cancer". Understanding Cancer. Stanford Medicine.
- "Lactate dehydrogenase test: MedlinePlus Medical Encyclopedia". MedlinePlus. U.S. National Library of Medicine.
- Butt AA, Michaels S, Greer D, Clark R, Kissinger P, Martin DH (July 2002). "Serum LDH level as a clue to the diagnosis of histoplasmosis". AIDS Read. 12 (7): 317–21. PMID 12161854.
- Heffner JE, Brown LK, Barbieri CA (April 1997). "Diagnostic value of tests that discriminate between exudative and transudative pleural effusions. Primary Study Investigators". Chest. 111 (4): 970–80. doi:10.1378/chest.111.4.970. PMID 9106577.
- Light RW, Macgregor MI, Luchsinger PC, Ball WC (October 1972). "Pleural effusions: the diagnostic separation of transudates and exudates". Ann. Intern. Med. 77 (4): 507–13. doi:10.7326/0003-4819-77-4-507. PMID 4642731.
- Joseph J, Badrinath P, Basran GS, Sahn SA (November 2001). "Is the pleural fluid transudate or exudate? A revisit of the diagnostic criteria". Thorax. 56 (11): 867–70. doi:10.1136/thorax.56.11.867. PMC 1745948. PMID 11641512.
- Joseph J, Badrinath P, Basran GS, Sahn SA (March 2002). "Is albumin gradient or fluid to serum albumin ratio better than the pleural fluid lactate dehydroginase in the diagnostic of separation of pleural effusion?". BMC Pulm Med. 2: 1. doi:10.1186/1471-2466-2-1. PMC 101409. PMID 11914151.
- Stein JH (1998). Internal Medicine. Elsevier Health Sciences. pp. 1408–. ISBN 978-0-8151-8698-4. Retrieved 12 August 2013.
- Warburg O (1956). "On the origin of cancer cells". Science. 123 (3191): 309–14. Bibcode:1956Sci...123..309W. doi:10.1126/science.123.3191.309. PMID 13298683.
- Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ (1997). "Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts". Science. 275 (5306): 1649–52. doi:10.1126/science.275.5306.1649. PMID 9054359. S2CID 19733670.
- Ramanathan R, Mancini RA, Suman SP, Beach CM (2014). "Covalent Binding of 4-Hydroxy-2-nonenal to Lactate Dehydrogenase Decreases NADH Formation and Metmyoglobin Reducing Activity". J Agric Food Chem. 62 (9): 2112–7. doi:10.1021/jf404900y. PMID 24552270.
- Xu HN, Kadlececk S, Profka H, Glickson JD, Rizi R, Li LZ (2014). "Is Higher Lactate an Indicator of Tumor Metastatic Risk? A Pilot MRS Study Using Hyperpolarized (13)C-Pyruvate". Acad Radiol. 21 (2): 223–31. doi:10.1016/j.acra.2013.11.014. PMC 4169113. PMID 24439336.
- Kim HS, Lee HE, Yang HK, Kim WH (2014). "High lactate dehydrogenase 5 expression correlates with high tumoral and stromal vascular endothelial growth factor expression in gastric cancer". Pathobiology. 81 (2): 78–85. doi:10.1159/000357017. PMID 24401755.
- Dym O, Pratt EA, Ho C, Eisenberg D (August 2000). "The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme". Proc. Natl. Acad. Sci. U.S.A. 97 (17): 9413–8. Bibcode:2000PNAS...97.9413D. doi:10.1073/pnas.97.17.9413. PMC 16878. PMID 10944213.
Further reading
- Johnson WT, Canfield WK (1985). "Intestinal absorption and excretion of zinc in streptozotocin-diabetic rats as affected by dietary zinc and protein". J Nutr. 115 (9): 1217–27. doi:10.1093/jn/115.9.1217. PMID 3897486.
- Ein SH, Mancer K, Adeyemi SD (1985). "Malignant sacrococcygeal teratoma--endodermal sinus, yolk sac tumor--in infants and children: a 32-year review". J Pediatr Surg. 20 (5): 473–7. doi:10.1016/s0022-3468(85)80468-1. PMID 3903096.
- Azuma M, Shi M, Danenberg KD, Gardner H, Barrett C, Jacques CJ, Sherod A, Iqbal S, El-Khoueiry A, Yang D, Zhang W, Danenberg PV, Lenz HJ (2007). "Serum lactate dehydrogenase levels and glycolysis significantly correlate with tumor VEGFA and VEGFR expression in metastatic CRC patients". Pharmacogenomics. 8 (12): 1705–13. doi:10.2217/14622416.8.12.1705. PMID 18086000.
- Masepohl B, Klipp W, Pühler A (1988). "Genetic characterization and sequence analysis of the duplicated nifA/nifB gene region of Rhodobacter capsulatus". Mol Gen Genet. 212 (1): 27–37. doi:10.1007/bf00322441. PMID 2836706. S2CID 21009965.
- Galardo MN, Regueira M, Riera MF, Pellizzari EH, Cigorraga SB, Meroni SB (2014). "Lactate Regulates Rat Male Germ Cell Function through Reactive Oxygen Species". PLOS ONE. 9 (1): e88024. Bibcode:2014PLoSO...988024G. doi:10.1371/journal.pone.0088024. PMC 3909278. PMID 24498241.
- Tesch P, Sjödin B, Thorstensson A, Karlsson J (1978). "Muscle fatigue and its relation to lactate accumulation and LDH activity in man". Acta Physiol Scand. 103 (4): 413–20. doi:10.1111/j.1748-1716.1978.tb06235.x. PMID 716962.
- Beck O, Jernström B, Martinez M, Repke DB (1988). "In vitro study of the aromatic hydroxylation of 1-methyltetrahydro-beta-carboline (methtryptoline) in rat". Chem Biol Interact. 65 (1): 97–106. doi:10.1016/0009-2797(88)90034-8. PMID 3345575.
- Summermatter S, Santos G, Pérez-Schindler J, Handschin C (2013). "Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A." Proc Natl Acad Sci U S A. 110 (21): 8738–43. Bibcode:2013PNAS..110.8738S. doi:10.1073/pnas.1212976110. PMC 3666691. PMID 23650363.
- Robergs RA, Ghiasvand F, Parker D (2004). "Biochemistry of exercise-induced metabolic acidosis". Am J Physiol Regul Integr Comp Physiol. 287 (3): R502-16. doi:10.1152/ajpregu.00114.2004. PMID 15308499.
- Kresge N, Simoni RD, Hill RL (2005). "Otto Fritz Meyerhof and the elucidation of the glycolytic pathway". J Biol Chem. 280 (4): e3. PMID 15665335.
External links
 Media related to Lactate dehydrogenase at Wikimedia Commons
Media related to Lactate dehydrogenase at Wikimedia Commons
_1I10.png.webp)