Telomerase RNA component
Telomerase RNA component, also known as TR, TER or TERC, is an ncRNA found in eukaryotes that is a component of telomerase, the enzyme used to extend telomeres.[2][3] TERC serves as a template for telomere replication (reverse transcription) by telomerase. Telomerase RNAs differ greatly in sequence and structure between vertebrates, ciliates and yeasts, but they share a 5' pseudoknot structure close to the template sequence. The vertebrate telomerase RNAs have a 3' H/ACA snoRNA-like domain.[4][5][6]
| TERC | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Aliases | TERC, DKCA1, PFBMFT2, SCARNA19, TR, TRC3, hTR, Telomerase RNA component | ||||||
| External IDs | OMIM: 602322 GeneCards: TERC | ||||||
| Orthologs | |||||||
| Species | Human | Mouse | |||||
| Entrez |
| ||||||
| Ensembl |
| ||||||
| UniProt |
|
| |||||
| RefSeq (mRNA) |
|
| |||||
| RefSeq (protein) |
|
| |||||
| Location (UCSC) | n/a | n/a | |||||
| PubMed search | [1] | n/a | |||||
| Wikidata | |||||||
| |||||||
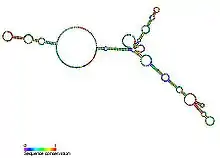
| Vertebrate telomerase RNA | |
|---|---|
 | |
| Identifiers | |
| Symbol | Telomerase-vert |
| Rfam | RF00024 |
| Other data | |
| RNA type | Gene |
| Domain(s) | Eukaryote; Virus |
| PDB structures | PDBe |
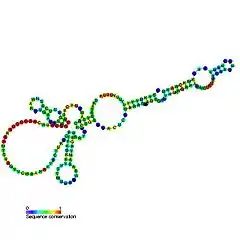
| Ciliate telomerase RNA | |
|---|---|
 | |
| Identifiers | |
| Symbol | Telomerase-cil |
| Rfam | RF00025 |
| Other data | |
| RNA type | Gene |
| Domain(s) | Eukaryote |
| PDB structures | PDBe |
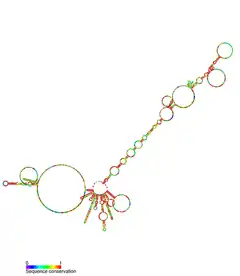
| Saccharomyces cerevisiae telomerase RNA | |
|---|---|
 | |
| Identifiers | |
| Symbol | Sacc_telomerase |
| Rfam | RF01050 |
| Other data | |
| RNA type | Gene |
| Domain(s) | Eukaryote |
| PDB structures | PDBe |
Structure
TERC is a Long non-coding RNA (lncRNA) ranging in length from ~150nt in ciliates to 400-600nt in vertebrates, and 1,300nt in yeast (Alnafakh). Mature human TERC (hTR) is 451nt in length.[7] TERC has extensive secondary structural features over 4 principal conserved domains.[8] The core domain, the largest domain at the 5’ end of TERC, contains the CUAAC Telomere template sequence. Its secondary structure consists of a large loop containing the template sequence, a P1 loop-closing helix, and a P2/P3 pseudoknot.[9] The core domain and CR4/CR5 conserved domain associate with TERT, and are the only domains of TERC necessary for in vitro catalytic activity of telomerase.[10] The 3’ end of TERC consists of a conserved H/ACA domain,[9] a 2 hairpin structure connected by a single-stranded hinge and bordered on the 3’ end by a single-stranded ACA sequence.[7] The H/ACA domain binds Dyskerin, GAR1, NOP10, NHP2, to form an H/ACA RNP complex.[9] The conserved CR7 domain is also localized at the 3’ end of TERC, and contains a 3nt CAB (Cajal body Localisation) box which binds TCAB1.[9]
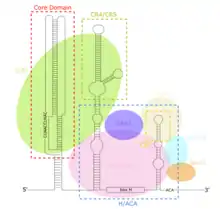
Function
Telomerase is a ribonucleoprotein polymerase that maintains telomere ends by addition of the telomere repeat TTAGGG. This repeat does vary across eukaryotes (see the table on the telomere article for a complete list). The enzyme consists of a protein component (TERT) with reverse transcriptase activity, and an RNA component, encoded by this gene, that serves as a template for the telomere repeat. CCCUAA found near position 50 of the vertebrate TERC sequence acts as the template. Telomerase expression plays a role in cellular senescence, as it is normally repressed in postnatal somatic cells resulting in progressive shortening of telomeres. Deregulation of telomerase expression in somatic cells may be involved in oncogenesis. Studies in mice suggest that telomerase also participates in chromosomal repair, since de novo synthesis of telomere repeats may occur at double-stranded breaks.[11] Homologs of TERC can also be found in the Gallid herpes viruses.[12]
The core domain of TERC contains the RNA template from which TERT synthesizes TTAGGG telomeric repeats.[9] Unlike in other RNPs, in telomerase, the protein TERT is catalytic while the lncRNA TERC is structural, rather than acting as a ribozyme.[13] The core region of TERC and TERT are sufficient to reconstitute catalytic telomerase activity in vitro.[9][10] The H/ACA domain of TERC recruits the Dyskerin complex (DKC1, GAR1, NOP10, NHP2), which stabilises TERC, increasing telomerase complex formation and overall catalytic activity.[9] The CR7 domain binds TCAB1, which localizes telomerase to cajal bodies, further increasing telomerase catalytic activity.[9] TERC is ubiquitously expressed, even in cells lacking telomerase activity and TERT expression.[14] As a result, various TERT-independent functional roles of TERC have been proposed. 14 genes containing a TERC binding motif are directly transcriptionally regulated by TERC through RNA-DNA triplex formation-mediated increase of expression. TERC-mediated upregulation of Lin37, Trpg1l, tyrobp, Usp16 stimulates the NF-κB pathway, resulting in increased expression and secretion of inflammatory cytokines.[15]
Biosynthesis
Unlike most lncRNAs which are assembled from introns by the spliceosome, hTR is directly transcribed from a dedicated promoter site[7] located at genomic locus 3q26.2[16] by RNA polymerase II.[7] Mature hTR is 451nt in length, but approximately 1/3 of cellular hTR transcripts at steady state have ~10nt genomically encoded 3’ tails. The majority of those extended hTR species have additional oligo-A 3’ extension.[7] Processing of immature 3’-tailed hTR to mature 451nt hTR can be accomplished by direct 3’-5’ exoribonucleolytic degradation or by an indirect pathway of oligoadenylation by PAPD5, removal of 3’ oligo-A tail by the 3’-5’ RNA exonuclease PARN, and subsequent 3’-5’ exoribonucleolytic degradation.[7] Extended hTR transcripts are also degraded by the RNA exosome.[7]
The 5’ ends of hTR transcripts are also additionally processed. TGS-1 hypermethylation the 5'-methylguanosine cap to an N2,2,7 trimethylguanosine (TMG) cap, which inhibits hTR maturation.[17] Binding of the Dyskerin complex to transcribed H/ACA domains of hTR during transcription promotes termination of transcription.[7] Control of the relative rates of these various competing pathways that activate or inhibit hTR maturation is a crucial element of regulation of overall telomerase activity.
Clinical Significance
Loss of function mutations in the TERC genomic locus have been associated with a variety of degenerative diseases. Mutations in TERC have been associated with dyskeratosis congenita,[18] idiopathic pulmonary fibrosis,[19] aplastic anemia, and myelodysplasia.[9] Overexpression and improper regulation of TERC have been associated with a variety of cancers. Upregulation of hTR is widely observed in patients with precancerous cervical phenotype as a result of HPV infection.[20] Overexpression of TERC enhances MDV-mediated oncogenesis,[21] and is observed in gastric carcinoma.[22] Overexpression of TERC is also observed in inflammatory conditions such as Type II diabetes and multiple sclerosis, due to TERC-mediated activation of the NF-κB inflammatory pathway.[15]
TERC has been implicated as protective in osteoporosis, with its increased expression arresting the rate of osteogenesis.[23] Due to its overexpression in a range of cancer phenotypes, TERC has been investigated as a potential cancer biomarker. It was found to be an effective biomarker of lung squamous cell carcinoma (LUSC).[24]
References
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, et al. (September 1995). "The RNA component of human telomerase". Science. 269 (5228): 1236–41. Bibcode:1995Sci...269.1236F. doi:10.1126/science.7544491. PMID 7544491. S2CID 9440710.
- Jády BE, Richard P, Bertrand E, Kiss T (February 2006). "Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres". Molecular Biology of the Cell. 17 (2): 944–54. doi:10.1091/mbc.E05-09-0904. PMC 1356602. PMID 16319170.
- McCormick-Graham M, Romero DP (April 1995). "Ciliate telomerase RNA structural features". Nucleic Acids Research. 23 (7): 1091–7. doi:10.1093/nar/23.7.1091. PMC 306816. PMID 7739888.
- Lingner J, Hendrick LL, Cech TR (August 1994). "Telomerase RNAs of different ciliates have a common secondary structure and a permuted template". Genes & Development. 8 (16): 1984–98. doi:10.1101/gad.8.16.1984. PMID 7958872.
- Theimer CA, Feigon J (June 2006). "Structure and function of telomerase RNA". Current Opinion in Structural Biology. 16 (3): 307–18. doi:10.1016/j.sbi.2006.05.005. PMID 16713250.
- Roake CM, Chen L, Chakravarthy AL, Ferrell JE, Raffa GD, Artandi SE (May 2019). "Disruption of Telomerase RNA Maturation Kinetics Precipitates Disease". Molecular Cell. 74 (4): 688–700.e3. doi:10.1016/j.molcel.2019.02.033. PMC 6525023. PMID 30930056.
- Alnafakh RA, Adishesh M, Button L, Saretzki G, Hapangama DK (2019). "Telomerase and Telomeres in Endometrial Cancer". Frontiers in Oncology. 9: 344. doi:10.3389/fonc.2019.00344. PMC 6533802. PMID 31157162.
- Zhang Q, Kim NK, Feigon J (December 2011). "Architecture of human telomerase RNA". Proceedings of the National Academy of Sciences of the United States of America. 108 (51): 20325–32. Bibcode:2011PNAS..10820325Z. doi:10.1073/pnas.1100279108. PMC 3251123. PMID 21844345.
- Webb CJ, Zakian VA (August 2016). "Telomerase RNA is more than a DNA template". RNA Biology. 13 (8): 683–9. doi:10.1080/15476286.2016.1191725. PMC 4993324. PMID 27245259.
- "Entrez Gene: TERC telomerase RNA component".
- Fragnet L, Kut E, Rasschaert D (June 2005). "Comparative functional study of the viral telomerase RNA based on natural mutations". The Journal of Biological Chemistry. 280 (25): 23502–15. doi:10.1074/jbc.M501163200. PMID 15811851. S2CID 24301693.
- Wang Y, Sušac L, Feigon J (December 2019). "Structural Biology of Telomerase". Cold Spring Harbor Perspectives in Biology. 11 (12): a032383. doi:10.1101/cshperspect.a032383. PMC 6886448. PMID 31451513.
- Shay JW, Wright WE (May 2019). "Telomeres and telomerase: three decades of progress". Nature Reviews Genetics. 20 (5): 299–309. doi:10.1038/s41576-019-0099-1. PMID 30760854. S2CID 61156603.
- Liu H, Yang Y, Ge Y, Liu J, Zhao Y (September 2019). "TERC promotes cellular inflammatory response independent of telomerase". Nucleic Acids Research. 47 (15): 8084–8095. doi:10.1093/nar/gkz584. PMC 6735767. PMID 31294790.
- "OMIM Entry - * 602322 - TELOMERASE RNA COMPONENT; TERC". www.omim.org. Retrieved 2020-03-02.
- Chen L, Roake CM, Galati A, Bavasso F, Micheli E, Saggio I, et al. (February 2020). "Loss of Human TGS1 Hypermethylase Promotes Increased Telomerase RNA and Telomere Elongation". Cell Reports. 30 (5): 1358–1372.e5. doi:10.1016/j.celrep.2020.01.004. PMC 7156301. PMID 32023455.
- Rich RR (2018-01-13). Clinical immunology : principles and practice (Fifth ed.). [St. Louis, Mo.] ISBN 978-0-7020-7039-6. OCLC 1023865227.
- Swigris JJ, Brown KK (2018-07-25). Idiopathic pulmonary fibrosis. St. Louis. ISBN 978-0-323-54432-0. OCLC 1053744041.
- Liu Y, Fan P, Yang Y, Xu C, Huang Y, Li D, et al. (November 2019). "Human papillomavirus and human telomerase RNA component gene in cervical cancer progression". Scientific Reports. 9 (1): 15926. Bibcode:2019NatSR...915926L. doi:10.1038/s41598-019-52195-5. PMC 6828729. PMID 31685833.
- Kheimar A, Trimpert J, Groenke N, Kaufer BB (March 2019). "Overexpression of cellular telomerase RNA enhances virus-induced cancer formation". Oncogene. 38 (10): 1778–1786. doi:10.1038/s41388-018-0544-1. PMID 30846849. S2CID 53085869.
- Heine B, Hummel M, Demel G, Stein H (June 1998). "Demonstration of constant upregulation of the telomerase RNA component in human gastric carcinomas using in situ hybridization". The Journal of Pathology. 185 (2): 139–44. doi:10.1002/(SICI)1096-9896(199806)185:2<139::AID-PATH79>3.0.CO;2-L. PMID 9713339.
- Gao GC, Yang DW, Liu W (January 2020). "LncRNA TERC alleviates the progression of osteoporosis by absorbing miRNA-217 to upregulate RUNX2". European Review for Medical and Pharmacological Sciences. 24 (2): 526–534. doi:10.26355/eurrev_202001_20029. PMID 32016954.
- Storti CB, de Oliveira RA, de Carvalho M, Hasimoto EN, Cataneo DC, Cataneo AJ, et al. (February 2020). "Telomere-associated genes and telomeric lncRNAs are biomarker candidates in lung squamous cell carcinoma (LUSC)". Experimental and Molecular Pathology. 112: 104354. doi:10.1016/j.yexmp.2019.104354. PMID 31837325.
Further reading
- de Lange T, Jacks T (August 1999). "For better or worse? Telomerase inhibition and cancer". Cell. 98 (3): 273–5. doi:10.1016/S0092-8674(00)81955-8. PMID 10458601. S2CID 14642341.
- Marrone A, Dokal I (December 2004). "Dyskeratosis congenita: molecular insights into telomerase function, ageing and cancer". Expert Reviews in Molecular Medicine. 6 (26): 1–23. doi:10.1017/S1462399404008671. PMID 15613268.
- Yamaguchi H (June 2007). "Mutations of telomerase complex genes linked to bone marrow failures". Journal of Nippon Medical School. 74 (3): 202–9. doi:10.1272/jnms.74.202. PMID 17625368.
- Zaug AJ, Linger J, Cech TR (February 1996). "Method for determining RNA 3' ends and application to human telomerase RNA". Nucleic Acids Research. 24 (3): 532–3. doi:10.1093/nar/24.3.532. PMC 145649. PMID 8602368.
- Soder AI, Hoare SF, Muire S, Balmain A, Parkinson EK, Keith WN (April 1997). "Mapping of the gene for the mouse telomerase RNA component, Terc, to chromosome 3 by fluorescence in situ hybridization and mouse chromosome painting". Genomics. 41 (2): 293–4. doi:10.1006/geno.1997.4621. PMID 9143511.
- Zhao JQ, Hoare SF, McFarlane R, Muir S, Parkinson EK, Black DM, Keith WN (March 1998). "Cloning and characterization of human and mouse telomerase RNA gene promoter sequences". Oncogene. 16 (10): 1345–50. doi:10.1038/sj.onc.1201892. PMID 9546436. S2CID 2699389.
- Mitchell JR, Wood E, Collins K (December 1999). "A telomerase component is defective in the human disease dyskeratosis congenita". Nature. 402 (6761): 551–5. Bibcode:1999Natur.402..551M. doi:10.1038/990141. PMID 10591218. S2CID 4430482.
- Chen JL, Blasco MA, Greider CW (March 2000). "Secondary structure of vertebrate telomerase RNA". Cell. 100 (5): 503–14. doi:10.1016/S0092-8674(00)80687-X. PMID 10721988. S2CID 15642776.
- Wong KK, Chang S, Weiler SR, Ganesan S, Chaudhuri J, Zhu C, et al. (September 2000). "Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation". Nature Genetics. 26 (1): 85–8. doi:10.1038/79232. PMID 10973255. S2CID 1873111.
- Mitchell JR, Collins K (August 2000). "Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase". Molecular Cell. 6 (2): 361–71. doi:10.1016/S1097-2765(00)00036-8. PMID 10983983.
- Imoto I, Pimkhaokham A, Fukuda Y, Yang ZQ, Shimada Y, Nomura N, et al. (August 2001). "SNO is a probable target for gene amplification at 3q26 in squamous-cell carcinomas of the esophagus". Biochemical and Biophysical Research Communications. 286 (3): 559–65. doi:10.1006/bbrc.2001.5428. PMID 11511096.
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I (September 2001). "The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita". Nature. 413 (6854): 432–5. Bibcode:2001Natur.413..432V. doi:10.1038/35096585. PMID 11574891. S2CID 4348062.
- Pruzan R, Pongracz K, Gietzen K, Wallweber G, Gryaznov S (January 2002). "Allosteric inhibitors of telomerase: oligonucleotide N3'-->P5' phosphoramidates". Nucleic Acids Research. 30 (2): 559–68. doi:10.1093/nar/30.2.559. PMC 99832. PMID 11788719.
- Zhang RG, Zhang RP, Wang XW, Xie H (March 2002). "Effects of cisplatin on telomerase activity and telomere length in BEL-7404 human hepatoma cells". Cell Research. 12 (1): 55–62. doi:10.1038/sj.cr.7290110. PMID 11942411. S2CID 36839452.
- Yang Y, Chen Y, Zhang C, Huang H, Weissman SM (July 2002). "Nucleolar localization of hTERT protein is associated with telomerase function". Experimental Cell Research. 277 (2): 201–9. doi:10.1006/excr.2002.5541. PMID 12083802.
- Chang JT, Chen YL, Yang HT, Chen CY, Cheng AJ (July 2002). "Differential regulation of telomerase activity by six telomerase subunits". European Journal of Biochemistry. 269 (14): 3442–50. doi:10.1046/j.1432-1033.2002.03025.x. PMID 12135483.
- Gavory G, Farrow M, Balasubramanian S (October 2002). "Minimum length requirement of the alignment domain of human telomerase RNA to sustain catalytic activity in vitro". Nucleic Acids Research. 30 (20): 4470–80. doi:10.1093/nar/gkf575. PMC 137139. PMID 12384594.
- Sood AK, Coffin J, Jabbari S, Buller RE, Hendrix MJ, Klingelhutz A (2003). "p53 null mutations are associated with a telomerase negative phenotype in ovarian carcinoma". Cancer Biology & Therapy. 1 (5): 511–7. doi:10.4161/cbt.1.5.167. PMID 12496479. S2CID 45381814.
- Antal M, Boros E, Solymosy F, Kiss T (February 2002). "Analysis of the structure of human telomerase RNA in vivo". Nucleic Acids Research. 30 (4): 912–20. doi:10.1093/nar/30.4.912. PMC 100349. PMID 11842102.
