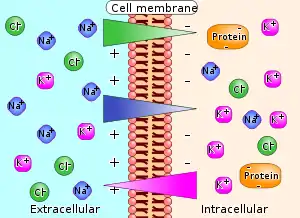
Electrochemical gradient
An electrochemical gradient is a gradient of electrochemical potential, usually for an ion that can move across a membrane. The gradient consists of two parts, the chemical gradient, or difference in solute concentration across a membrane, and the electrical gradient, or difference in charge across a membrane. When there are unequal concentrations of an ion across a permeable membrane, the ion will move across the membrane from the area of higher concentration to the area of lower concentration through simple diffusion. Ions also carry an electric charge that forms an electric potential across a membrane. If there is an unequal distribution of charges across the membrane, then the difference in electric potential generates a force that drives ion diffusion until the charges are balanced on both sides of the membrane.[1]
Definition
The electrochemical gradient is the gradient of the electrochemical potential:
- , with
- the chemical potential of the ion species
- the valency of the ion species
- F, Faraday constant
- the local electric potential
Overview
Electrochemical potential is important in electroanalytical chemistry and industrial applications such as batteries and fuel cells. It represents one of the many interchangeable forms of potential energy through which energy may be conserved.
In biological processes, the direction an ion moves by diffusion or active transport across a membrane is determined by the electrochemical gradient. In mitochondria and chloroplasts, proton gradients are used to generate a chemiosmotic potential that is also known as a proton motive force. This potential energy is used for the synthesis of ATP by oxidative phosphorylation or photophosphorylation, respectively.[2]
An electrochemical gradient has two components. First, the electrical component is caused by a charge difference across the lipid membrane. Second, a chemical component is caused by a differential concentration of ions across the membrane. The combination of these two factors determines the thermodynamically favourable direction for an ion's movement across a membrane.[1][3]
An electrochemical gradient is analogous to the water pressure across a hydroelectric dam. Membrane transport proteins such as the sodium-potassium pump within the membrane are equivalent to turbines that convert the water's potential energy to other forms of physical or chemical energy, and the ions that pass through the membrane are equivalent to water that ends up at the bottom of the dam. Also, energy can be used to pump water up into the lake above the dam. In similar manner, chemical energy in cells can be used to create electrochemical gradients.[4][5]
Chemistry
The term is typically applied in contexts wherein a chemical reaction is to take place, such as one involving the transfer of an electron at a battery electrode. In a battery, an electrochemical potential arising from the movement of ions balances the reaction energy of the electrodes. The maximum voltage that a battery reaction can produce is sometimes called the standard electrochemical potential of that reaction (see also Electrode potential and Table of standard electrode potentials). In instances pertaining specifically to the movement of electrically charged solutes, the potential is often expressed in units of volts. See: Concentration cell.
Biological context
The generation of a transmembrane electrical potential through ion movement across a cell membrane drives biological processes like nerve conduction, muscle contraction, hormone secretion, and sensory processes. By convention, a typical animal cell has a transmembrane electrical potential of -50 mV to -70 mV inside the cell relative to the outside.[6]
Electrochemical gradients also play a role in establishing proton gradients in oxidative phosphorylation in mitochondria. The final step of cellular respiration is the electron transport chain. Four complexes embedded in the inner membrane of the mitochondrion make up the electron transport chain. However, only complexes I, III, and IV pump protons from the matrix to the intermembrane space (IMS). In total, there are ten protons translocated from the matrix to the IMS which generates an electrochemical potential of more than 200mV. This drives the flux of protons back into the matrix through ATP synthase which produces ATP by adding an inorganic phosphate to ADP.[7] Thus, generation of a proton electrochemical gradient is crucial for energy production in mitochondria.[8] The total equation for the electron transport chain is:
NADH + 11 H+(matrix) + 1/2 O2 → NAD+ + 10 H+(IMS) + H2O.[9]
Similar to the electron transport chain, the light-dependent reactions of photosynthesis pump protons into the thylakoid lumen of chloroplasts to drive the synthesis of ATP by ATP synthase. The proton gradient can be generated through either noncyclic or cyclic photophosphorylation. Of the proteins that participate in noncyclic photophosphorylation, photosystem II (PSII), plastiquinone, and cytochrome b6f complex directly contribute to generating the proton gradient. For each four photons absorbed by PSII, eight protons are pumped into the lumen.[10] The total equation for photophosphorylation is:
2 NADP+ + 6 H+(stroma) + 2 H2O → 2 NADPH + 8 H+(lumen) + O2.[11]
Several other transporters and ion channels play a role in generating a proton electrochemical gradient. One is TPK3, a potassium channel that is activated by Ca2+ and conducts K+ from the thylakoid lumen to the stroma which helps establish the pH gradient. On the other hand, the electro-neutral K+ efflux antiporter (KEA3) transports K+ into the thylakoid lumen and H+ into the stroma which helps establish the electric field.[12]
Ion gradients

Since the ions are charged, they cannot pass through the membrane via simple diffusion. Two different mechanisms can transport the ions across the membrane: active or passive transport. An example of active transport of ions is the Na+-K+-ATPase (NKA). NKA catalyzes the hydrolysis of ATP into ADP and an inorganic phosphate and for every molecule of ATP hydrolized, three Na+ are transported outside and two K+ are transported inside the cell. This makes the inside of the cell more negative than the outside and more specifically generates a membrane potential Vmembrane of about -60mV.[5] An example of passive transport is ion fluxes through Na+, K+, Ca2+, and Cl− channels. These ions tend to move down their concentration gradient. For example, since there is a high concentration of Na+ outside the cell, Na+ will flow through the Na+ channel into the cell. Since the electric potential inside the cell is negative, the influx of a positive ion depolarizes the membrane which brings the transmembrane electric potential closer to zero. However, Na+ will continue moving down its concentration gradient as long as the effect of the chemical gradient is greater than the effect of the electrical gradient. Once the effect of both gradients are equal (for Na+ this at a membrane potential of about +70mV), the influx of Na+ stops because the driving force (ΔG) is zero. The equation for the driving force is:[13][14]
In this equation, R represents the gas constant, T represents absolute temperature, z is the ionic charge, and F represents the Faraday constant.[15]
Cellular ion concentrations are given in the table below. X- represents proteins with a net negative charge.
| Ion | Mammal | Squid axon | S. cerevisiae | E. coli | Sea water | ||
|---|---|---|---|---|---|---|---|
| Cell | Blood | Cell | Blood | ||||
| K+ | 100 - 140 | 4-5 | 400 | 10 - 20 | 300 | 30 - 300 | 10 |
| Na+ | 5-15 | 145 | 50 | 440 | 30 | 10 | 500 |
| Mg2+ | 10 [lower-alpha 1] 0.5 - 0.8 [lower-alpha 2] |
1 - 1.5 | 50 | 30 - 100 [lower-alpha 1] 0.01 - 1 [lower-alpha 2] |
50 | ||
| Ca2+ | 10−4 | 2.2 - 2.6 [lower-alpha 3] 1.3 - 1.5 [lower-alpha 4] |
10−4 - 3×10−4 | 10 | 2 | 3 [lower-alpha 1] 10−4 [lower-alpha 2] |
10 |
| Cl− | 4 | 110 | 40 - 150 | 560 | 10 - 200 [lower-alpha 5] | 500 | |
| X− | 138 | 9 | 300 - 400 | 5-10 | |||
| HCO3− | 12 | 29 | |||||
| pH | 7.1 - 7.3[20] | 7.35 to 7.45 [20] (normal arterial blood pH)
6.9 - 7.8 [20] (overall range) |
7.2 - 7.8[21] | 8.1 - 8.2[22] | |||
- Bound
- Free
- Total
- Ionised
- Medium dependent
Proton gradients
Proton gradients in particular are important in many types of cells as a form of energy storage. The gradient is usually used to drive ATP synthase, flagellar rotation, or transport of metabolites.[23] This section will focus on three processes that help establish proton gradients in their respective cells: bacteriorhodopsin and noncyclic photophosphorylation and oxidative phosphorylation.
Bacteriorhodopsin
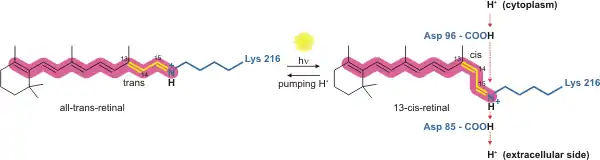
The way bacteriorhodopsin generates a proton gradient in Archaea is through a proton pump. The proton pump relies on proton carriers to drive protons from the side of the membrane with a low H+ concentration to the side of the membrane with a high H+ concentration. In bacteriorhodopsin, the proton pump is activated by absorption of photons of 568 nm wavelength which leads to isomerization of the Schiff base (SB) in retinal forming the K state. This moves SB away from Asp85 and Asp212, causing H+ transfer from the SB to Asp85 forming the M1 state. The protein then shifts to the M2 state by separating Glu204 from Glu194 which releases a proton from Glu204 into the external medium. The SB is reprotonated by Asp96 which forms the N state. It is important that the second proton comes from Asp96 since its deprotonated state is unstable and rapidly reprotonated with a proton from the cytosol. The protonation of Asp85 and Asp96 causing re-isomerization of the SB forming the O state. Finally, bacteriorhodopsin returns to its resting state when Asp85 releases its proton to Glu204.[23][24]
Photophosphorylation
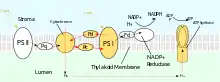
PSII also relies on light to drive the formation of proton gradients in chloroplasts, however PSII utilizes vectorial redox chemistry to achieve this goal. Rather than physically transporting protons through the protein, reactions requiring the binding of protons will occur on the extracellular side while reactions requiring the release of protons will occur on the intracellular side. Absorption of photons of 680 nm wavelength is used to excite two electrons in P680 to a higher energy level. These higher energy electrons are transferred to protein-bound plastoquinone (PQA) and then to unbound plastoquinone (PQB). This reduces plastoquinone (PQ) to plastoquinol (PQH2) which is released from PSII after gaining two protons from the stroma. The electrons in P680 are replenished by oxidizing water through the oxygen-evolving complex (OEC). This results in release of O2 and H+ into the lumen.[23] The total reaction is shown:
After being released from PSII, PQH2 travels to the cytochrome b6f complex which then transfers two electrons from PQH2 to plastocyanin in two separate reactions. The process that occurs is similar to the Q-cycle in Complex III of the electron transport chain. In the first reaction, PQH2 binds to the complex on the lumen side and one electron is transferred to the iron-sulfur center which then transfers it to cytochrome f which then transfers it to plastocyanin. The second electron is transferred to heme bL which then transfers it to heme bH which then transfers it to PQ. In the second reaction, a second PQH2 gets oxidized, adding an electron to another plastocyanin and PQ. Both reactions together transfer four protons into the lumen.[25][26]
Oxidative phosphorylation
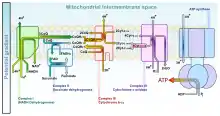
In the electron transport chain, complex I (CI) catalyzes the reduction of ubiquinone (UQ) to ubiquinol (UQH2) by the transfer of two electrons from reduced nicotinamide adenine dinucleotide (NADH) which translocates four protons from the mitochondrial matrix to the IMS:[27]
Complex III (CIII) catalyzes the Q-cycle. The first step involving the transfer of two electrons from the UQH2 reduced by CI to two molecules of oxidized cytochrome c at the Qo site. In the second step, two more electrons reduce UQ to UQH2 at the Qi site.[27] The total reaction is shown:
Complex IV (CIV) catalyzes the transfer of two electrons from the cytochrome c reduced by CIII to one half of a full oxygen. Utilizing one full oxygen in oxidative phosphorylation requires the transfer of four electrons. The oxygen will then consume four protons from the matrix to form water while another four protons are pumped into the IMS.[27] The total reaction is shown:
See also
References
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. p. 403. ISBN 978-1-4292-3414-6.
- Nath, Sunil; Villadsen, John (2015-03-01). "Oxidative phosphorylation revisited". Biotechnology and Bioengineering. 112 (3): 429–437. doi:10.1002/bit.25492. ISSN 1097-0290. PMID 25384602. S2CID 2598635.
- Yang, Huanghe; Zhang, Guohui; Cui, Jianmin (2015-01-01). "BK channels: multiple sensors, one activation gate". Frontiers in Physiology. 6: 29. doi:10.3389/fphys.2015.00029. PMC 4319557. PMID 25705194.
- Shattock, Michael J.; Ottolia, Michela; Bers, Donald M.; Blaustein, Mordecai P.; Boguslavskyi, Andrii; Bossuyt, Julie; Bridge, John H. B.; Chen-Izu, Ye; Clancy, Colleen E. (2015-03-15). "Na+/Ca2+ exchange and Na+/K+-ATPase in the heart". The Journal of Physiology. 593 (6): 1361–1382. doi:10.1113/jphysiol.2014.282319. ISSN 1469-7793. PMC 4376416. PMID 25772291.
- Aperia, Anita; Akkuratov, Evgeny E.; Fontana, Jacopo Maria; Brismar, Hjalmar (2016-04-01). "Na+-K+-ATPase, a new class of plasma membrane receptors". American Journal of Physiology. Cell Physiology. 310 (7): C491–C495. doi:10.1152/ajpcell.00359.2015. ISSN 0363-6143. PMID 26791490.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. p. 464. ISBN 978-1-4292-3414-6.
- Poburko, Damon; Demaurex, Nicolas (2012-04-24). "Regulation of the mitochondrial proton gradient by cytosolic Ca2+ signals" (PDF). Pflügers Archiv: European Journal of Physiology. 464 (1): 19–26. doi:10.1007/s00424-012-1106-y. ISSN 0031-6768. PMID 22526460. S2CID 18133149.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. pp. 743–745. ISBN 978-1-4292-3414-6.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. p. 744. ISBN 978-1-4292-3414-6.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. pp. 769–770. ISBN 978-1-4292-3414-6.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. p. 770. ISBN 978-1-4292-3414-6.
- Höhner, Ricarda; Aboukila, Ali; Kunz, Hans-Henning; Venema, Kees (2016-01-01). "Proton Gradients and Proton-Dependent Transport Processes in the Chloroplast". Frontiers in Plant Science. 7: 218. doi:10.3389/fpls.2016.00218. PMC 4770017. PMID 26973667.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. pp. 464–465. ISBN 978-1-4292-3414-6.
- Eisenberg, Bob (2013-05-07). "Interacting Ions in Biophysics: Real is not Ideal". Biophysical Journal. 104 (9): 1849–1866. arXiv:1305.2086. Bibcode:2013BpJ...104.1849E. doi:10.1016/j.bpj.2013.03.049. PMC 3647150. PMID 23663828.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. p. 465. ISBN 978-1-4292-3414-6.
- Philips, Ron Milo & Ron. "» What are the concentrations of different ions in cells?". Retrieved 2019-06-07.
- Lodish, Harvey; Berk, Arnold; Zipursky, S. Lawrence; Matsudaira, Paul; Baltimore, David; Darnell, James (2000). "Table 15-1, Typical Ion Concentrations in Invertebrates and Vertebrates". www.ncbi.nlm.nih.gov. Retrieved 2019-06-07.
- "The following table gives an idea of the intra and extra cellular ion concentrations in a squid axon and a mammalian cell". www.chm.bris.ac.uk. Retrieved 2019-06-07.
- Diem K, Lenter C. Scientific Tables. 565 (Seventh ed.). Basel: Ciba-Geigy Limited. pp. 653–654. ISBN 978-3-9801244-0-9.
- Spitzer, Kenneth W.; Vaughan-Jones, Richard D. (2003), Karmazyn, Morris; Avkiran, Metin; Fliegel, Larry (eds.), "Regulation of Intracellular pH in Mammalian Cells", The Sodium-Hydrogen Exchanger: From Molecule to its Role in Disease, Springer US, pp. 1–15, doi:10.1007/978-1-4615-0427-6_1, ISBN 9781461504276
- Slonczewski, Joan L.; Wilks, Jessica C. (2007-08-01). "pH of the Cytoplasm and Periplasm of Escherichia coli: Rapid Measurement by Green Fluorescent Protein Fluorimetry". Journal of Bacteriology. 189 (15): 5601–5607. doi:10.1128/JB.00615-07. ISSN 0021-9193. PMC 1951819. PMID 17545292.
- Brewer, Peter G. (September 1, 2008). "Rising Acidity in the Ocean: The Other CO2 Problem". doi:10.1038/scientificamericanearth0908-22. Cite journal requires
|journal=(help) - Gunner, M. R.; Amin, Muhamed; Zhu, Xuyu; Lu, Jianxun (2013-08-01). "Molecular mechanisms for generating transmembrane proton gradients". Biochimica et Biophysica Acta (BBA) - Bioenergetics. Metals in Bioenergetics and Biomimetics Systems. 1827 (8–9): 892–913. doi:10.1016/j.bbabio.2013.03.001. PMC 3714358. PMID 23507617.
- Wickstrand, Cecilia; Dods, Robert; Royant, Antoine; Neutze, Richard (2015-03-01). "Bacteriorhodopsin: Would the real structural intermediates please stand up?". Biochimica et Biophysica Acta (BBA) - General Subjects. Structural biochemistry and biophysics of membrane proteins. 1850 (3): 536–553. doi:10.1016/j.bbagen.2014.05.021. PMID 24918316.
- Nelson, David; Cox, Michael (2013). Lehninger Principles of Biochemistry. New York: W.H. Freeman. pp. 782–783. ISBN 978-1-4292-3414-6.
- Schöttler, Mark Aurel; Tóth, Szilvia Z.; Boulouis, Alix; Kahlau, Sabine (2015-05-01). "Photosynthetic complex stoichiometry dynamics in higher plants: biogenesis, function, and turnover of ATP synthase and the cytochrome b 6 f complex". Journal of Experimental Botany. 66 (9): 2373–2400. doi:10.1093/jxb/eru495. ISSN 0022-0957. PMID 25540437.
- Sun, Fei; Zhou, Qiangjun; Pang, Xiaoyun; Xu, Yingzhi; Rao, Zihe (2013-08-01). "Revealing various coupling of electron transfer and proton pumping in mitochondrial respiratory chain". Current Opinion in Structural Biology. 23 (4): 526–538. doi:10.1016/j.sbi.2013.06.013. PMID 23867107.
- Campbell & Reece (2005). Biology. Pearson Benjamin Cummings. ISBN 978-0-8053-7146-8.
- Stephen T. Abedon, "Important words and concepts from Chapter 8, Campbell & Reece, 2002 (1/14/2005)", for Biology 113 at the Ohio State University