Evolution of biological complexity
The evolution of biological complexity is one important outcome of the process of evolution.[1] Evolution has produced some remarkably complex organisms – although the actual level of complexity is very hard to define or measure accurately in biology, with properties such as gene content, the number of cell types or morphology all proposed as possible metrics.[2][3][4]
Many biologists used to believe that evolution was progressive (orthogenesis) and had a direction that led towards so-called "higher organisms", despite a lack of evidence for this viewpoint.[5] This idea of "progression" and "higher organisms" in evolution is now regarded as misleading, with natural selection having no intrinsic direction and organisms selected for either increased or decreased complexity in response to local environmental conditions.[6] Although there has been an increase in the maximum level of complexity over the history of life, there has always been a large majority of small and simple organisms and the most common level of complexity appears to have remained relatively constant.
Selection for simplicity and complexity
Usually organisms that have a higher rate of reproduction than their competitors have an evolutionary advantage. Consequently, organisms can evolve to become simpler and thus multiply faster and produce more offspring, as they require fewer resources to reproduce. A good example are parasites such as Plasmodium – the parasite responsible for malaria – and mycoplasma; these organisms often dispense with traits that are made unnecessary through parasitism on a host.[7]
A lineage can also dispense with complexity when a particular complex trait merely provides no selective advantage in a particular environment. Loss of this trait need not necessarily confer a selective advantage, but may be lost due to the accumulation of mutations if its loss does not confer an immediate selective disadvantage.[8] For example, a parasitic organism may dispense with the synthetic pathway of a metabolite where it can readily scavenge that metabolite from its host. Discarding this synthesis may not necessarily allow the parasite to conserve significant energy or resources and grow faster, but the loss may be fixed in the population through mutation accumulation if no disadvantage is incurred by loss of that pathway. Mutations causing loss of a complex trait occur more often than mutations causing gain of a complex trait.
With selection, evolution can also produce more complex organisms. Complexity often arises in the co-evolution of hosts and pathogens,[9] with each side developing ever more sophisticated adaptations, such as the immune system and the many techniques pathogens have developed to evade it. For example, the parasite Trypanosoma brucei, which causes sleeping sickness, has evolved so many copies of its major surface antigen that about 10% of its genome is devoted to different versions of this one gene. This tremendous complexity allows the parasite to constantly change its surface and thus evade the immune system through antigenic variation.[10]
More generally, the growth of complexity may be driven by the co-evolution between an organism and the ecosystem of predators, prey and parasites to which it tries to stay adapted: as any of these become more complex in order to cope better with the diversity of threats offered by the ecosystem formed by the others, the others too will have to adapt by becoming more complex, thus triggering an ongoing evolutionary arms race[9] towards more complexity.[11] This trend may be reinforced by the fact that ecosystems themselves tend to become more complex over time, as species diversity increases, together with the linkages or dependencies between species.
Types of trends in complexity
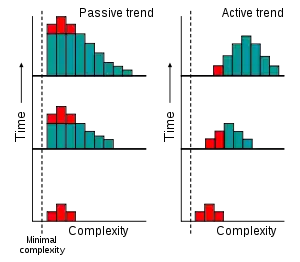
If evolution possessed an active trend toward complexity (orthogenesis), as was widely believed in the 19th century,[12] then we would expect to see an active trend of increase over time in the most common value (the mode) of complexity among organisms.[13]
However, an increase in complexity can also be explained through a passive process.[13] Assuming unbiased random changes of complexity and the existence of a minimum complexity leads to an increase over time of the average complexity of the biosphere. This involves an increase in variance, but the mode does not change. The trend towards the creation of some organisms with higher complexity over time exists, but it involves increasingly small percentages of living things.[4]
In this hypothesis, any appearance of evolution acting with an intrinsic direction towards increasingly complex organisms is a result of people concentrating on the small number of large, complex organisms that inhabit the right-hand tail of the complexity distribution and ignoring simpler and much more common organisms. This passive model predicts that the majority of species are microscopic prokaryotes, which is supported by estimates of 106 to 109 extant prokaryotes[14] compared to diversity estimates of 106 to 3·106 for eukaryotes.[15][16] Consequently, in this view, microscopic life dominates Earth, and large organisms only appear more diverse due to sampling bias.
Genome complexity has generally increased since the beginning of the life on Earth.[17][18] Some computer models have suggested that the generation of complex organisms is an inescapable feature of evolution.[19][20] Proteins tend to become more hydrophobic over time,[21] and to have their hydrophobic amino acids more interspersed along the primary sequence.[22] Increases in body size over time are sometimes seen in what is known as Cope's rule.[23]
Constructive neutral evolution
Recently work in evolution theory has proposed that by relaxing selection pressure, which typically acts to streamline genomes, the complexity of an organism increases by a process called constructive neutral evolution.[24] Since the effective population size in eukaryotes (especially multi-cellular organisms) is much smaller than in prokaryotes,[25] they experience lower selection constraints.
According to this model, new genes are created by non-adaptive processes, such as by random gene duplication. These novel entities, although not required for viability, do give the organism excess capacity that can facilitate the mutational decay of functional subunits. If this decay results in a situation where all of the genes are now required, the organism has been trapped in a new state where the number of genes has increased. This process has been sometimes described as a complexifying ratchet.[26] These supplemental genes can then be co-opted by natural selection by a process called neofunctionalization. In other instances constructive neutral evolution does not promote the creation of new parts, but rather promotes novel interactions between existing players, which then take on new moonlighting roles.[26]
Constructive neutral evolution has also been used to explain how ancient complexes, such as the spliceosome and the ribosome, have gained new subunits over time, how new alternative spliced isoforms of genes arise, how gene scrambling in ciliates evolved, how pervasive pan-RNA editing may have arisen in Trypanosoma brucei, how functional lncRNAs have likely arisen from transcriptional noise, and how even useless protein complexes can persist for millions of years.[24][27][26][28][29][30][31]
History
In the 19th century, some scientists such as Jean-Baptiste Lamarck (1744–1829) and Ray Lankester (1847–1929) believed that nature had an innate striving to become more complex with evolution. This belief may reflect then-current ideas of Hegel (1770–1831) and of Herbert Spencer (1820–1903) which envisaged the universe gradually evolving to a higher, more perfect state.
This view regarded the evolution of parasites from independent organisms to a parasitic species as "devolution" or "degeneration", and contrary to nature. Social theorists have sometimes interpreted this approach metaphorically to decry certain categories of people as "degenerate parasites". Later scientists regarded biological devolution as nonsense; rather, lineages become simpler or more complicated according to whatever forms had a selective advantage.[32]
See also
References
- Werner, Andreas; Piatek, Monica J.; Mattick, John S. (April 2015). "Transpositional shuffling and quality control in male germ cells to enhance evolution of complex organisms". Annals of the New York Academy of Sciences. 1341 (1): 156–163. Bibcode:2015NYASA1341..156W. doi:10.1111/nyas.12608. PMC 4390386. PMID 25557795.
- Adami, C. (2002). "What is complexity?". BioEssays. 24 (12): 1085–94. doi:10.1002/bies.10192. PMID 12447974.
- Waldrop, M.; et al. (2008). "Language: Disputed definitions". Nature. 455 (7216): 1023–1028. doi:10.1038/4551023a. PMID 18948925.
- Longo, Giuseppe; Montévil, Maël (2012-01-01). Dinneen, Michael J.; Khoussainov, Bakhadyr; Nies, André (eds.). Computation, Physics and Beyond. Lecture Notes in Computer Science. Springer Berlin Heidelberg. pp. 289–308. CiteSeerX 10.1.1.640.1835. doi:10.1007/978-3-642-27654-5_22. ISBN 9783642276538.
- McShea, D. (1991). "Complexity and evolution: What everybody knows". Biology and Philosophy. 6 (3): 303–324. doi:10.1007/BF00132234. S2CID 53459994.
- Ayala, F. J. (2007). "Darwin's greatest discovery: design without designer". PNAS. 104 (Suppl 1): 8567–73. Bibcode:2007PNAS..104.8567A. doi:10.1073/pnas.0701072104. PMC 1876431. PMID 17494753.
- Sirand-Pugnet, P.; Lartigue, C.; Marenda, M.; et al. (2007). "Being Pathogenic, Plastic, and Sexual while Living with a Nearly Minimal Bacterial Genome". PLOS Genet. 3 (5): e75. doi:10.1371/journal.pgen.0030075. PMC 1868952. PMID 17511520.
- Maughan, H.; Masel, J.; Birky, W. C.; Nicholson, W. L. (2007). "The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis". Genetics. 177 (2): 937–948. doi:10.1534/genetics.107.075663. PMC 2034656. PMID 17720926.
- Dawkins, Richard; Krebs, J. R. (1979). "Arms Races between and within Species". Proceedings of the Royal Society B. 205 (1161): 489–511. Bibcode:1979RSPSB.205..489D. doi:10.1098/rspb.1979.0081. PMID 42057. S2CID 9695900.
- Pays, E. (2005). "Regulation of antigen gene expression in Trypanosoma brucei". Trends Parasitol. 21 (11): 517–20. doi:10.1016/j.pt.2005.08.016. PMID 16126458.
- Heylighen, F. (1999a) "The Growth of Structural and Functional Complexity during Evolution", in F. Heylighen, J. Bollen & A. Riegler (eds.) The Evolution of Complexity Kluwer Academic, Dordrecht, 17–44.
- Ruse, Michael (1996). Monad to man: the Concept of Progress in Evolutionary Biology. Harvard University Press. pp. 526–529 and passim. ISBN 978-0-674-03248-4.
- Carroll SB (2001). "Chance and necessity: the evolution of morphological complexity and diversity". Nature. 409 (6823): 1102–9. Bibcode:2001Natur.409.1102C. doi:10.1038/35059227. PMID 11234024. S2CID 4319886.
- Oren, A. (2004). "Prokaryote diversity and taxonomy: current status and future challenges". Philos. Trans. R. Soc. Lond. B Biol. Sci. 359 (1444): 623–38. doi:10.1098/rstb.2003.1458. PMC 1693353. PMID 15253349.
- May, R. M.; Beverton, R. J. H. (1990). "How Many Species?". Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 330 (1257): 293–304. doi:10.1098/rstb.1990.0200.
- Schloss, P.; Handelsman, J. (2004). "Status of the microbial census". Microbiol Mol Biol Rev. 68 (4): 686–91. doi:10.1128/MMBR.68.4.686-691.2004. PMC 539005. PMID 15590780.
- Markov, A. V.; Anisimov, V. A.; Korotayev, A. V. (2010). "Relationship between genome size and organismal complexity in the lineage leading from prokaryotes to mammals". Paleontological Journal. 44 (4): 363–373. doi:10.1134/s0031030110040015. S2CID 10830340.
- Sharov, Alexei A (2006). "Genome increase as a clock for the origin and evolution of life". Biology Direct. 1 (1): 17. doi:10.1186/1745-6150-1-17. PMC 1526419. PMID 16768805.
- Furusawa, C.; Kaneko, K. (2000). "Origin of complexity in multicellular organisms". Phys. Rev. Lett. 84 (26 Pt 1): 6130–3. arXiv:nlin/0009008. Bibcode:2000PhRvL..84.6130F. doi:10.1103/PhysRevLett.84.6130. PMID 10991141. S2CID 13985096.
- Adami, C.; Ofria, C.; Collier, T. C. (2000). "Evolution of biological complexity". PNAS. 97 (9): 4463–8. arXiv:physics/0005074. Bibcode:2000PNAS...97.4463A. doi:10.1073/pnas.97.9.4463. PMC 18257. PMID 10781045.
- Wilson, Benjamin A.; Foy, Scott G.; Neme, Rafik; Masel, Joanna (24 April 2017). "Young genes are highly disordered as predicted by the preadaptation hypothesis of de novo gene birth". Nature Ecology & Evolution. 1 (6): 0146–146. doi:10.1038/s41559-017-0146. PMC 5476217. PMID 28642936.
- Foy, Scott G.; Wilson, Benjamin A.; Bertram, Jason; Cordes, Matthew H. J.; Masel, Joanna (April 2019). "A Shift in Aggregation Avoidance Strategy Marks a Long-Term Direction to Protein Evolution". Genetics. 211 (4): 1345–1355. doi:10.1534/genetics.118.301719. PMC 6456324. PMID 30692195.
- Heim, N. A.; Knope, M. L.; Schaal, E. K.; Wang, S. C.; Payne, J. L. (2015-02-20). "Cope's rule in the evolution of marine animals". Science. 347 (6224): 867–870. Bibcode:2015Sci...347..867H. doi:10.1126/science.1260065. PMID 25700517.
- Stoltzfus, Arlin (1999). "On the Possibility of Constructive Neutral Evolution". Journal of Molecular Evolution. 49 (2): 169–181. Bibcode:1999JMolE..49..169S. CiteSeerX 10.1.1.466.5042. doi:10.1007/PL00006540. ISSN 0022-2844. PMID 10441669. S2CID 1743092.
- Sung, W.; Ackerman, M. S.; Miller, S. F.; Doak, T. G.; Lynch, M. (2012). "Drift-barrier hypothesis and mutation-rate evolution". Proceedings of the National Academy of Sciences. 109 (45): 18488–18492. Bibcode:2012PNAS..10918488S. doi:10.1073/pnas.1216223109. PMC 3494944. PMID 23077252.
- Lukeš, Julius; Archibald, John M.; Keeling, Patrick J.; Doolittle, W. Ford; Gray, Michael W. (2011). "How a neutral evolutionary ratchet can build cellular complexity". IUBMB Life. 63 (7): 528–537. doi:10.1002/iub.489. PMID 21698757. S2CID 7306575.
- Gray, M. W.; Lukes, J.; Archibald, J. M.; Keeling, P. J.; Doolittle, W. F. (2010). "Irremediable Complexity?". Science. 330 (6006): 920–921. Bibcode:2010Sci...330..920G. doi:10.1126/science.1198594. ISSN 0036-8075. PMID 21071654. S2CID 206530279.
- Daniel, Chammiran; Behm, Mikaela; Öhman, Marie (2015). "The role of Alu elements in the cis-regulation of RNA processing". Cellular and Molecular Life Sciences. 72 (21): 4063–4076. doi:10.1007/s00018-015-1990-3. ISSN 1420-682X. PMID 26223268. S2CID 17960570.
- Covello, PatrickS.; Gray, MichaelW. (1993). "On the evolution of RNA editing". Trends in Genetics. 9 (8): 265–268. doi:10.1016/0168-9525(93)90011-6. PMID 8379005.
- Palazzo, Alexander F.; Koonin, Eugene V. (2020). "Functional Long Non-coding RNAs Evolve from Junk Transcripts". Cell. 183 (5): 1151–1161. doi:10.1016/j.cell.2020.09.047. ISSN 0092-8674.
- Hochberg, GKA; Liu, Y; Marklund, EG; Metzger, BPH; Laganowsky, A; Thornton, JW (December 2020). "A hydrophobic ratchet entrenches molecular complexes". Nature. 588 (7838): 503–508. doi:10.1038/s41586-020-3021-2. PMID 33299178.
- Dougherty, Michael J. (July 1998). "Is the human race evolving or devolving?". Scientific American.
From a biological perspective, there is no such thing as devolution. All changes in the gene frequencies of populations—and quite often in the traits those genes influence—are by definition evolutionary changes. [...] When species do evolve, it is not out of need but rather because their populations contain organisms with variants of traits that offer a reproductive advantage in a changing environment.

