Evolution of the wolf
The evolution of the wolf occurred over a geologic time scale of at least 300 thousand years. The grey wolf Canis lupus is a highly adaptable species that is able to exist in a range of environments and which possesses a wide distribution across the Holarctic. Studies of modern grey wolves have identified distinct sub-populations that live in close proximity to each other.[2][3] This variation in sub-populations is closely linked to differences in habitat – precipitation, temperature, vegetation, and prey specialization – which affect cranio-dental plasticity.[4][5][6][7]
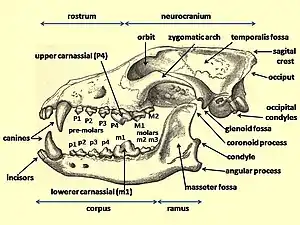
_Wolf_Cranium.png.webp)
The archaeological and paleontological records show grey wolf continuous presence for at least the last 300,000 years.[8] This continuous presence contrasts with genomic analyses, which suggest that all modern wolves and dogs descend from a common ancestral wolf population[9][10][11] that existed as recently as 20,000 years ago.[9] These analyses indicate a population bottleneck, followed by a rapid radiation from an ancestral population at a time during, or just after, the Last Glacial Maximum. This implies that the original wolf populations were out-competed by a new type of wolf which replaced them. However, the geographic origin of this radiation is not known.
Fossil record

The fossil record for ancient vertebrates is composed of rarely occurring fragments from which it is often impossible to obtain genetic material. Researchers are limited to morphologic analysis but it is difficult to estimate the intra-species and inter-species variations and relationships that existed between specimens across time and place. Some observations are debated by researchers who do not always agree, and hypotheses that are supported by some authors are challenged by others.[12]
There is general agreement on the most ancient record, which shows that feliforms and caniforms emerged within the super-family Carnivoramorpha 43 million years before present (YBP).[13] The caniforms included the fox-like genus Leptocyon whose various species existed from 34 million YBP before branching 11.9 million YBP into Vulpes (foxes) and Canini (canines). The jackal-sized Eucyon existed in North America from 10 million YBP and by the Early Pliocene about 6–5 million YBP the coyote-like Eucyon davisi[14] invaded Eurasia. In North America it gave rise to early Canis which first appeared in the Miocene (6 million YBP) in south-western US and Mexico. By 5 million YBP the larger Canis lepophagus appeared in the same region.[15]:p58
The canids that had immigrated from North America to Eurasia – Eucyon, Vulpes, and Nyctereutes – were small to medium-sized predators during the Late Miocene and Early Pliocene but they were not the top predators. The position of the canids would change with the arrival of Canis to become a dominant predator across the Holarctic. The wolf-sized C. chihliensis appeared in northern China in the Mid-Pliocene around 4–3 million YBP.[15]:p148 The large wolf-sized Canis appeared in the Middle Pliocene about 3 million years ago in the Yushe Basin, Shanxi Province, China. By 2.5 million years ago its range included the Nihewan Basin in Yangyuan County, Hebei, China and Kuruksay, Tadzhikistan.[16] This was followed by an explosion of Canis evolution across Eurasia in the Early Pleistocene around 1.8 million YBP in what is commonly referred to as the wolf event. It is associated with the formation of the mammoth steppe and continental glaciation. Canis spread to Europe in the forms of C. arnensis, C. etruscus, and C. falconeri.[15]:p148
The fossil record is incomplete but it is likely that wolves arose from a population of small, early canids.[17]:p241 Morphological evidence[17]:p239[18] and genetic evidence[19] both suggest that wolves evolved during the Pliocene and Early Pleistocene eras from the same lineage that also led to the coyote,[17]:p239 with fossil specimens indicating that the coyote and the wolf diverged from a common ancestor 1.5 million years ago.[17]:p240[18] The ancestor of the jackal and the other extant members of the genus Canis had split from the lineage before this time.[17]:p240
After this separation from a common ancestor the species that were believed to be involved in the further evolution of the wolf and coyote – and the beliefs of some paleontologists – diverged.[17]:p240 A number of researchers believed that the lines of C. priscolatrans, C. etruscus, C. rufus and C. lupus were components involved in some way that lead to the modern wolf and coyote.[17]:p240[20][21][22][23][24][25]
| Wolf evolution | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Proposed evolution and branching of genus Canis towards the wolf.[17]:p240 |
Canis lepophagus
Canis lepophagus lived in the early Pliocene in North America.[15] Kurten proposed that the Blancan C. lepophagus[26] derived from smaller Miocene Canis species in North America. It then became widespread across Eurasia where it was either identical to, or closely related with, C. arnensis of Europe.[17]:p241[27][28]
Johnston describes C. lepophagus as having a more slender skull and skeleton than in the modern coyote.[29]:385 Robert M. Nowak found that the early populations had small, delicate and narrowly proportioned skulls that resemble small coyotes and appear to be ancestral to C. latrans.[17]:p241 Johnson noted that some specimens found in Cita Canyon, Texas, had larger, broader skulls,[29] and along with other fragments Nowak suggested that these were evolving into wolves.[17]:p241[18]
Tedford disagreed with previous authors and found that its cranio-dental morphology lacked some characteristics that are shared by C. lupus and C. latrans, and therefore there was not a close relationship but it did suggest C. lepophagus was the ancestor of both wolves and coyotes.[30]:p119
Canis priscolatrans
Canis priscolatrans lived in the late Pliocene-Early Pleistocene in North America.[18] The first definite wolf appeared in the Late Blancan/Early Irvingtonian,[17]:p240[18][31] and named C. priscolatrans that was either very close to[27][28] or a synonym for Canis edwardii.[17]:p241[18]:82[32][33] It resembled C. rufus in cranial size and proportions but with more complex dentition.[17]:p241 However, there are no fossils of C. rufus until the Late Rancholabrean.[17]:p242
Kurtén was uncertain if C. priscolatrans derived from C. lepophagus and C. arnensis,[28] but believed that C. priscolatrans was a population of large coyotes that were ancestral to Rancholabrean and recent C. latrans. He noted that C. arnensis of Europe showed striking similarities to C. priscolatrans, and they could represent what once was a holarctic population of coyotes.[27]:p27 Nowak disagreed, and believed that C. priscolatrans was a counterpart to the European C. etruscus.[18] Kurtén later proposed that both C. priscolatrans and C. etruscus were part of a group which led to C. lupus but was not sure if they evolved separately from C. lepophagus or a possible common ancestor that was derived from C. lepophagus.[28]
The remains of the larger coyote-like Canis edwardii have been found in the later Pliocene in the south-western USA along with C. lepophagus, which indicates a descent.[15]:p60 Tedford recognised C. edwardii[34] and found that the cranio-dental morphology of C. priscolatrans fell inside that of C. edwardii such that the species name C. priscolatrans was doubtful (nomen dubium).[30]:p131
Canis armbrusteri
The North American wolves became larger, with tooth specimens indicating that C. priscolatrans diverged into the large wolf C. armbrusteri.[17]:p242[35] during the Middle Pleistocene in North America.[18] Robert A. Martin disagreed, and believed that C. armbrusteri[36] was C. lupus.[23] Nowak disagreed with Martin and proposed that C. armbrusteri was not related to C. lupus but C. priscolatrans, which then gave rise to C. dirus. Tedford proposed that the South American C. gezi and C. nehringi share dental and cranial similarities developed for hypercarnivory, suggesting C. armbrusteri was the common ancestor of C. gezi, C. nehringi and C. dirus.[30]:148
Canis dirus
In 1908 the paleontologist John Campbell Merriam began retrieving numerous fossilized bone fragments of a large wolf from the Rancho La Brea tar pits. By 1912 he had found a skeleton sufficiently complete to be able to formally recognize these and the previously found specimens under the name C. dirus (Leidy 1858).
Canis dirus[37] lived in the late Pleistocene to early Holocene in North and South America [38] and was the largest of all Canis species.[15]:52 In 1987, new hypothesis proposed that a mammal population could give rise to a larger form called a hypermorph during times when food was abundant, but when food later became scarce the hypermorph would either adapt to a smaller form or go extinct. This hypothesis might explain the large body sizes found in many Late Pleistocene mammals compared to their modern counterparts. Both extinction and speciation – a new species splitting from an older one – could occur together during periods of climatic extremes.[39][40] Gloria D. Goulet agreed with Martin and further proposed that this hypothesis might explain the sudden appearance of C. dirus in North America, and that because of the similarities in their skull shapes that C. lupus gave rise to the C. dirus hypermorph due to abundant game, a stable environment, and large competitors.[41] Nowak, Kurten and Berta disagreed with Goulet and proposed that C. dirus was not derived from C. lupus.[18][28][42] The three noted paleontologists Xiaoming Wang, R. H. Tedford and R. M. Nowak have all proposed that C. dirus had evolved from C. armbrusteri,[15]:p52[30]:181 with Nowak stating that there were specimens from Cumberland Cave, Maryland, that indicated C. armbrusteri diverging into C. dirus.[17]:p243[43] The two taxa share a number of characteristics (synapomorphy), which suggests an origin of C. dirus in the late Irvingtonian in the open terrain in the midcontinent, and then later expanding eastward and displacing its ancestor C. armbrusteri.[30]:181
dirus–lupus hybrids
Merriam named 3 unusual species based on specimens recovered from the Rancho La Brea tar pits. They were regarded by Nowak as taxonomic synonyms for Canis lupus.[18]
Canis occidentalis furlongi (Merriam 1910)[44] was described as a wolf considerably smaller than the dire wolf and more closely related to the timber wolf Canis lupus occidentalis. However, its premolar P4 (upper carnassials) were massive, and the hypocone of the molar M1 was larger than that of the dire wolf. One specimen's teeth and palate are described as being between the dire wolf and Canis lupus occidentalis.[44] Nowak proposed the name Canis lupus furlongi as he believed that it was a subspecies of the gray wolf.[18] There is the possibility that wolves living in marginal areas led to dire wolf/gray wolf hybrids.[45]
Canis milleri (Merriam 1912),[46] the Miller wolf, was as large as the timber wolf but with a shorter and heavier head. Its skull and dentition were described as being intermediate between Canis lupus occidentalis and the dire wolf. Its skull differed from occidentalis due to its wider skull, especially at the palate, and the size of its P4 and M1 were much larger than any known timber wolf, with the P4 approaching that of the dire wolf in size.[47] It is regarded by Nowak as a taxonomic synonym of Canis lupus furlongi.[18]
Aenocyon milleri (Merriam 1918)[48] was described as a wolf different from the dire wolf by its smaller size, low sagittal crest, and a less prominent inion, but closer to the dire wolf than the timber wolf. Only one specimen was found. It is regarded by Nowak as a taxonomic synonym of Canis lupus furlongi.[18]
Canis mosbachensis

Canis mosbachensis, sometimes known as the Mosbach wolf, is an extinct small wolf that once inhabited Eurasia from the Middle to Late Pleistocene.[49] The phylogenetic descent of the extant wolf C. lupus from C. etruscus through C. mosbachensis is widely accepted.[49][17]:239–245 In 2010, a study found that the diversity of the Canis group decreased by the end of the Early Pleistocene to Middle Pleistocene and was limited in Eurasia to the small wolves of the C. mosbachensis–C. variabilis group that were a comparable size to the extant Indian wolf (Canis lupus pallipes), and the large hypercarnivorous Canis (Xenocyon) lycaonoides that was comparable in size to extant northern grey wolves.[8]
Canis variabilis
In 2012, a study of the wolf-like Canis species of ancient China under the direction of Xiaoming Wang found that these were all quite close to C. lupus in both dental and post-cranial dimensions except for Canis variabilis, which was "very strange" compared to other Canis in China as it had much smaller cranio-dental dimensions than earlier and later species.[50] The study concluded that "It is very likely that this species is the ancestor of the domestic dog Canis familiaris, a hypothesis that has been proposed by previous authors."[51][52][53][54][55]
Canis chihliensis
| Wolf evolution – alternate proposal | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Proposed evolution and branching from Eucyon towards the wolf.[15]:p148[30]:p181 |
Wang and Tedford proposed that the genus Canis was the descendant of the coyote-like Eucyon davisi, and its remains first appeared in the Miocene (6 million YBP) in south-western USA and Mexico. By the Pliocene (5 million YBP), the larger Canis lepophagus appeared in the same region and by the Early Pleistocene (1 million YBP) Canis latrans (the coyote) was in existence. They proposed that the progression from Eucyon davisi to C lepophagus to the coyote was linear evolution.[15]:p58 Additionally, C. edwardii, C. latrans and C. aureus form together a small clade and because C. edwardii appeared earliest spanning the mid-Blancan (late Pliocene) to the close of the Irvingtonian (late Pleistocene) it is proposed as the ancestor.[30]:p175,180
Nowak and Tedford also believed that it was possible for C. lupus to have been derived from a Miocene or Pliocene canid line that preceded and was separate from C. lepophagus.[18][31] Based on morphology from China, the Pliocene wolf C. chihliensis may have been the ancestor for both C. armbrusteri and C. lupus before their migration into North America.[15]:p148[30]:p181 C. chihliensis appears to be more primitive and smaller than C. lupus, and measurements of its skull and teeth are similar to C. lupus but those of its postcranial elements are smaller.[50] C. armbrusteri appeared in North America in the Middle Pleistocene and is a wolf-like form larger than any Canis at that time.[18] At the end of the most recent glacial retreat during the past 30,000 years, warming melted the glacial barriers across northern Canada allowing arctic mammals to extend their range into mid-latitude North America, including elk, caribou, bison, and the gray wolf.[15]:p61
In Eurasia during the Middle Pleistocene, C. falconeri gave rise to the hypercarnivore genus Xenocyon, which then gave rise to genus Cuon (the dhole) and genus Lycaon (the African hunting dog).[15]:p105,149 Just before the appearance of C. dirus, North America was invaded by genus Xenocyon that was as large as C. dirus and more hypercarnivorous. The fossil record shows them as rare and it is assumed that they could not compete with the newly derived C. dirus.[15]:p60
Canis antonii
The large wolf C. antonii from late Pliocene to early Pleistocene China was assessed as being a variation within C. chihliensis,[30]:p197 and the large wolf C. falconeri occurred abruptly in Europe in the Early Pleistocene, perhaps representing a westward extension of C. antonii.[30]:p181
Canis lupus
The earliest Canis lupus specimen was a fossil tooth discovered at Old Crow, Yukon, Canada. The specimen was found in sediment dated 1 million YBP,[30] however the geological attribution of this sediment is questioned.[30][56] Slightly younger specimens were discovered at Cripple Creek Sump, Fairbanks, Alaska, in strata dated 810,000 YBP. Both discoveries point to an origin of these wolves in east Beringia during the Middle Pleistocene.[30]
In France, the subspecies C. l. lunellensis Bonifay, 1971[57] discovered at Lunel-Viel, Hérault dated 400–350,000 YBP, C. l. santenaisiensis Argant, 1991[58] from Santenay, Côte-d'Or dated to 200,000 YBP, and C. lupus maximus Boudadi-Maligne, 2012[59] from Jaurens cave, Nespouls, Corrèze dated 31,000 YBP, show a progressive increase in size and are proposed to be chrono-subspecies.[60][12] In Italy, the earliest Canis lupus specimens were found at La Polledrara di Cecanibbio, 20 km north-west of Rome in strata dated 340,000–320,000 YBP.[61] In 2017, a study found that the dimensions of the upper and lower carnassial teeth of the early Holocene Italian wolf are close to those of C. l. maximus. Fluctuations in the size of C. lupus carnassial teeth correlate with the spread of megafauna. The Italian wolf underwent a reduction in body size with the loss of the red deer in Italy during the Renaissance.[12] The proposed lineage is:
C. etruscus → C. mosbachensis → C. l. lunellensis → C. l. santenaisiensis → C. l. maximus → C. l. lupus[12]
In 2020, a new species of wolf C. borjgali was discovered in Georgia. This wolf is proposed to be the next evolutionary step after C. mosbachensis and an ancestor of the C. lupus lineage.[62]
Canis c.f. familiaris (Paleolithic "dog")
There are a number of recently discovered specimens which are proposed as being Paleolithic dogs, however their taxonomy is debated. These have been found in either Europe or Siberia and date 40,000-17,000 YBP. They include Hohle Fels in Germany, Goyet Caves in Belgium, Predmosti in the Czech Republic, and four sites in Russia: Razboinichya Cave, Kostyonki-8, Ulakhan Sular, and Eliseevichi 1. Paw-prints from Chauvet Cave in France dated 26,000 YBP are suggested as being those of a dog, however these have been challenged as being left by a wolf.[63] Paleolithic dogs were directly associated with human hunting camps in Europe over 30,000 (YBP) and it is proposed that they were domesticated. They are also proposed to be either a proto-dog and the ancestor of the domestic dog or a type of wolf unknown to science.[64]
Canis lupus familiaris (domestic dog)
In 2002, a study was undertaken into the fossil skulls of two large canids that had been found buried within meters of the doorway of what was once a mammoth-bone hut at the Eliseevichi-I Upper Paleolithic site in the Bryansk Region on the Russian Plain, and using an accepted morphologically based definition of domestication declared them to be "Ice Age dogs". The carbon dating gave a calendar-year age estimate that ranged between 16,945–13,905 YBP.[65] In 2013, a study looked at one of these skulls and its mitochondrial DNA sequence was identified as Canis lupus familiaris.[66]
In 2015, a zooarchaeologist stated that "In terms of phenotypes, dogs and wolves are fundamentally different animals."[67]
In 1986, a study of skull morphology found that the domestic dog is morphologically distinct from all other canids except the wolf-like canids. "The difference in size and proportion between some breeds are as great as those between any wild genera, but all dogs are clearly members of the same species."[68] In 2010, a study of dog skull shape compared to extant carnivorans proposed that "The greatest shape distances between dog breeds clearly surpass the maximum divergence between species in the Carnivora. Moreover, domestic dogs occupy a range of novel shapes outside the domain of wild carnivorans."[69]
The domestic dog compared to the wolf shows the greatest variation in the size and shape of the skull (Evans 1979) that range from 7 to 28 cm in length (McGreevy 2004). Wolves are dolichocephalic (long skulled) but not as extreme as some breeds of such as greyhounds and Russian wolfhounds (McGreevy 2004). Canine brachycephaly (short-skulledness) is found only in domestic dogs and is related to paedomorphosis (Goodwin 1997). Puppies are born with short snouts, with the longer skull of dolichocephalic dogs emerging in later development (Coppinger 1995). Other differences in head shape between brachycephalic and dolichocephalic dogs include changes in the craniofacial angle (angle between the basilar axis and hard palate) (Regodón 1993), morphology of the temporomandibular joint (Dickie 2001), and radiographic anatomy of the cribriform plate (Schwarz 2000).[70]
Nowak indicated that orbital angle of the eye socket is an important characteristic defining the difference between the dog and the wolf, with the wolf having the lower angle. Nowak compared the orbital angles of four North American canines (including the Indian dog) and produced the following values in degrees: coyote-42.8, wolf-42.8, dog-52.9 dire wolf-53.1. The orbital angle of the eye socket was clearly larger in the dog than in the coyote and the wolf; why it was almost the same as that of the dire wolf was not commented on.[18]
Many authors have concluded that compared to the adult extant wolf, the adult domestic dog has a relatively reduced rostrum (front part of the skull), an elevated frontal bone, a wider palate, a broader cranium, and smaller teeth (Hildebrand1954; Clutton-Brock, Corbet & Hills 1976; Olsen 1985; Wayne 1986; Hemmer 1990; Morey 1990). Other authors have disagreed and have stated that these traits can overlap and vary within the two (Crockford 1999; Harrison 1973). Wolf cubs have similar relative skull proportions as adult dogs and this was proposed as evidence that the domestic dog is a neotenic wolf. This was proposed to be due to either human selection for juvenile appearance or due to a pleiotropic effect as a result of selection for juvenile behavior (Clutton-Brock 1977; Belyaev 1979; Wayne 1986; Coppinger and Schneider 1995). Wayne (1986) concluded that his dog samples did not have significant relative shortening of the rostrum compared to wolves, calling this identification feature into question.[51] A 2004 study that used 310 wolf skulls and over 700 dog skulls representing 100 breeds concluded that the evolution of dog skulls can generally not be described by heterochronic processes such as neoteny although some pedomorphic dog breeds have skulls that resemble the skulls of juvenile wolves.[71] "Dogs are not paedomorphic wolves."[72]
Compared to the wolf, dog dentition is relatively less robust (Olsen 1985; Hemmer 1990), which is proposed to be due to the relaxation of natural selection when wolves became commensal scavengers, or to artificial selection (Olsen 1985; Clutton-Brock 1995). However, Kieser and Groeneveld (1992) compared the mandibulo-dental measurements of jackals (C. adustus, C. mesomelas) and Cape foxes (Vulpes chama) to equivalent-sized dogs and found that the canines of these other canids tended to be slightly smaller and their second molars larger compared to dogs, otherwise the proportions were essentially the same in all species. They concluded that "...the teeth of canids appear to have evolved in concert with one another and relatively independently of differences in dimorphism, size or functional demands". This calls into question the assumption that dog teeth are relatively small due to recent selection, suggesting that dog dentition is plesiomorphic from an ancestor that was smaller than the wolf.[51]
The reduced body size of the early dog compared to a wolf is thought due to niche selection (Olsen 1985; Morey 1992; Coppinger & Coppinger 2001). Morey (1992:199) states that "Results...are consistent with a hypothesis that early domestic dogs are evolutionary paedomorphs, products of strong selection for ontogenetically channeled size reduction and alterations of reproductive timing associated with the new domestic way of life."[51] However, in an domestication experiment the domesticated foxes remained the same size as unselected foxes (Trutt 1999:167).[67]
Wayne (1986) concluded that the dog is closer in skull morphology to C. latrans, C. aureus, C. adustus, C. mesomelas, Cuon alpinus and Lycaon pictus than to the wolf. Dahr (1942) concluded that the shape of the dog brain case is closer to that of the coyote than to that of the wolf. Manwell and Baker (1983) reviewed Dahr's work with the addition of dental data for canids and concluded that the dog ancestor was probably within the range of 13.6–20.5 kg, which is smaller than the range 27–54 kg for extant wolves (Mech 1970) and is comparable with the Dingo.[51]
The auditory bulla of the dog is relatively smaller and flatter than that of the wolf (Harrison 1973; Clutton-Brock, Corbet & Hill 1976; Nowak 1979; Olsen 1985; Wayne 1986), which is proposed to be due to relaxed selection under domestication as the dog no longer required the acute hearing of the wolf. However, bulla shape has been shown to facilitate increased sensitivity to specific frequencies but shape and size may not be correlated with acuity (Ewer 1973). Therefore, the observed difference could be that the dog bulla has retained its ancestral shape.[51]
The ventral edge of the dog's horizontal ramus of the mandible has a convex curve that does not exist in the wolf (Olsen 1985; Clutton-Brock 1995), and no discussion of this difference could be found in the literature. However, Biknevicius and Van Valkenburgh (1997) noticed that the horizontal ramus of bone-processing predators is thicker dorso-ventrally at the point caudal to the site of bone processing. This thickening may have been a function for niche adaptation by the dog's ancestor.[51]
A description of the superficial brain morphology of jackals (C. mesomelas, C. aureus), coyotes (C. latrans), wolves (C. lupus, C. rufus), and dogs indicated that the cerebellum of the dog closely approximates that of the coyote, which is closely aligned with the jackals, and that the wolves show numerous brain traits distinct from the other species (Atkins and Dillon 1971). Wolves also have serological and biochemical traits distinct from dogs (Leone and Wiens 1956; Lauer, Kuyt & Baker 1969).[51]
During the Last Glacial Maximum, there was greater wolf genetic diversity than there is today,[9][66] and within the Pleistocene gray wolf population the variations between local environments would have encouraged a range of wolf ecotypes that were genetically, morphologically and ecologically distinct from one another.[73] One author has proposed that the most likely explanation for the different morphological characteristics of the dog compared to the wolf is that the dog's ancestor was adapted to a different niche than the wolf.[51]
Genetic record
DNA sequences
The mitochondria within each cell contain many copies of a small circular DNA genome and in mammals it is 16,000–18,000 base pairs in length. A cell contains hundreds or thousands of mitochondria and therefore the genes contained within those mitochondria are more abundant than the genes that occur in the nucleus of the cell.[74][75] The abundance of mitochondrial DNA (mDNA) is useful for the genetic analysis of ancient remains where the DNA has degraded.[75][76]
Mitochondrial DNA sequences have a higher mutation rate than the mutation rate of nuclear genes and for mammals this rate is 5–10 times faster.[75][77][78] The mitochondrial protein-coding genes evolve much faster and are powerful markers for inferring evolution history at category levels such as families, genera, and species. However, they have evolved at a faster rate than other DNA markers and there is a timing difference in its molecular clock that needs to be validated against other sources. The taxonomic status of uncertain species is better resolved through using nuclear DNA from the nucleus of the cell, which is more suitable for analyzing the recent history.[79] In most cases, mDNA is inherited from the maternal ancestor.[75][80] Therefore, phylogenetic analysis of mDNA sequences within species provides a history of maternal lineages that can be represented as a phylogenetic tree.[75][81][82]
- From specimen to phylogenetic tree
 The 33,500 year old skull of the "Altai dog"
The 33,500 year old skull of the "Altai dog"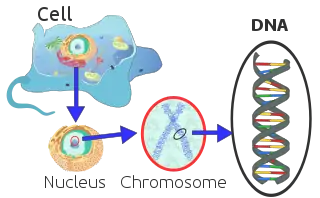
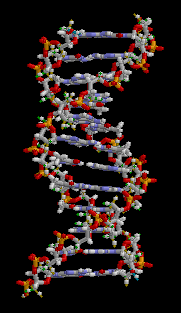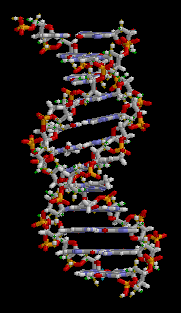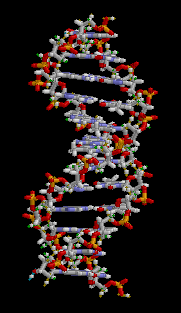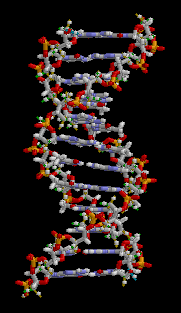 The structure of part of a DNA double helix
The structure of part of a DNA double helix
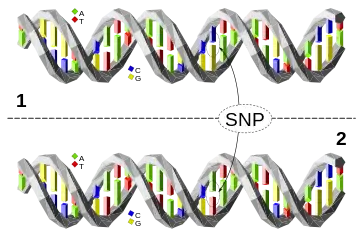 DNA molecule 1 differs from DNA molecule 2 at a single base pair location, called a single-nucleotide polymorphism (a SNP mutation)
DNA molecule 1 differs from DNA molecule 2 at a single base pair location, called a single-nucleotide polymorphism (a SNP mutation)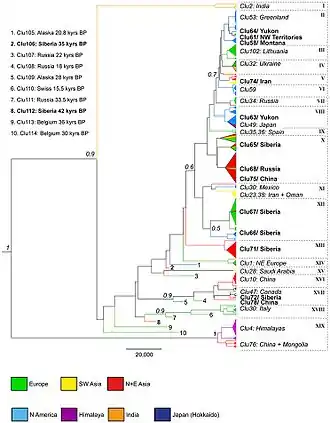 Phylogenetic tree of wolves under study
Phylogenetic tree of wolves under study
| Phylogenetic tree | |||||||||||||||||||||
| |||||||||||||||||||||
| Phylogenetic relationship between four canids.[83][84] |
The mutations that are different in these 4 sequences have been numbered and bolded. These mutations can then be used to build a phylogenetic tree for the four canids. In this example, the dog and gray wolf differ by two substitutions (highlighted in red), and each of them differs from the coyote by four substitutions.[75]
1 2 3 4 5 67
Golden Jackal A-G-C-T-G-T-C-GA-T-TC-CA
Coyote A-G-C-T-A-T-C-GA-A-TC-GA
Wolf T-G-C-T-A-T-G-GA-T-TC-CT
Dog T-G-G-T-A-T-G-GA-T-TC-CA
The mDNA sequences of the dog and wolf differ by only 0–12 substitutions within 261 base-pairs, whereas dogs always differed from coyotes and jackals by at least 20 substitutions.[75][85] This finding implies that the dog derived from the wolf and that there has been repeated back-crossing,[85] or that the dog may have descended from a now extinct species of canid whose closest living relative is the modern wolf.[86]
Marker issue
Different DNA studies may give conflicting results because of the specimens selected, the technology used, and the assumptions made by the researchers.[87] Any one from a panel of genetic markers can be chosen for use in a study. The techniques used to extract, locate and compare genetic sequences can be applied using advances in technology, which allows researchers to observe longer lengths of base pairs that provide more data to give better phylogenetic resolution.[88] Phylogenetic trees compiled using different genetic markers have given conflicting results on the relationship between the wolf, dog and coyote. One study based on SNPs[89] (a single mutation), and another based on nuclear gene sequences[90] (taken from the cell nucleus), showed dogs clustering with coyotes and separate from wolves. Another study based on SNPS showed wolves clustering with coyotes and separate from dogs.[91] Other studies based on a number of markers show the more widely accepted result of wolves clustering with dogs separate from coyotes.[85][92] These results demonstrate that caution is needed when interpreting the results provided by genetic markers.[89]
Timing issue
There are two key assumptions that are made for dating the divergence time for species: the generation time and the genetic mutation rate per generation. The time between generations for wolves is assumed to be three years based on the extant gray wolf, and two years for the dog based on the extant dog.[83] One recent major study assumed a generation time of 2 years for the dog for as far back as 10,000 years ago, and then assumed a generation time of 3 years (the same as the wolf) before that to calculate a proposed divergence time between the two.[9] In 2017, the wolf research scientist L. David Mech queried why evolutionary biologists were calculating the approximate time of the dog diverging from the wolf through using a wolf generation time of three years when published works using large data sets demonstrate a figure of 4.2–4.7 years. They were encouraged to recalculate their divergence dates accordingly.[93]
DNA studies are conducted but with "the mutation rate as the dominant source of uncertainty."[9] In 2005, Lindblad-Toh sequenced the first draft genome of the extant dog, and calculated a proposed mutation rate of 1x10−8 mutations per generation.[83] In 2015, Skoglund was able to sequence the first draft genome of the 35,000 YBP Taimyr wolf and used its radio-carbon date to validate a proposed genetic mutation rate of 0.4x10−8 mutations per generation.[10] The difference is a timing factor of 2.5, however another study stated that because only one Pleistocene wolf specimen has so far been sequenced, then the result should be treated with caution, with that study then providing both estimates to calculate the proposed divergence times between the wolf and dog.[11] However, in 2016 the mutation rate of the 4,800 YBP Newgrange dog matched that of the Taimyr wolf.[94]
Wolf-like canids
The wolf-like canids (the canid subfamily Caninae) are a group of large carnivores that are genetically closely related because their chromosomes number 78. The group includes genus Canis, Cuon and Lycaon. The members are the dog (C. lupus familiaris), gray wolf (C. lupus), coyote (C. latrans), golden jackal (C. aureus), Ethiopian wolf (C. simensis), black-backed jackal (Lupulella mesomelas), side-striped jackal (Lupulella adusta), dhole (Cuon alpinus), and African wild dog (Lycaon pictus).[95][96][97] Newly proposed members include the red wolf (Canis rufus), eastern wolf (Canis lycaon), and African golden wolf (C. anthus). As they possess 78 chromosomes, all members of the genus Canis (coyotes, wolves, jackals) are karyologically indistinguishable from each other, and from the dhole and the African hunting dog.[75]:p279[98] The members of Canis can potentially interbreed[86] and there is evidence that the Ethiopian wolf has hybridized with dogs.[99] According to zoologist Reginald Pocock, a dhole interbred with a golden jackal.[100] The African hunting dog is large, highly mobile, known to disperse over large distances and are rare throughout much of their geographical range,[101] making opportunities for hybridization difficult. A study of the maternal mitochondrial DNA of the black-backed jackal could find no evidence of genotypes from the most likely mates – the side-striped jackal nor the golden jackal – indicating that male black-backed jackals had not bred with these.[102] A search of the scientific literature could not find evidence of hybridization for the rare side-striped jackal.
A DNA sequence alignment for the wolf-like canids gave a phylogenetic tree with the gray wolf and dog being the most closely related, followed by a close affiliation with the coyote, golden jackal and Ethiopian wolf, and the dog can hybridize in the wild with these three species. Next closest to this group are the dhole and African wild dog that both have unique meat-slicing teeth, suggesting that this adaptation was later lost by the other members.[83] The two African jackals are shown as the most basal members of this clade, which means that this tree is indicating an African origin for the clade.[83][103] The tree illustrates the genotype-phenotype distinction, where a genotype is an organism's full hereditary information and a phenotype is an organism's actual observed properties, such as morphology, development, or behavior. By phenotype, the dhole (genus Cuon) and the African hunting dog (genus Lycaon) are not classified as members of the genus Canis, but by genotype they are closer to dogs, wolves and coyotes than are the two genus Canis jackals – the Side-striped jackal (C. adustus) and the Black-backed jackal (C. mesomelas).
In 2015, a study of mitochondrial genome sequences and nuclear genome sequences of African and Eurasian canids indicated that extant wolf-like canids had colonized Africa from Eurasia at least 5 times throughout the Pliocene and Pleistocene, which is consistent with fossil evidence suggesting that much of the African canid diversity resulted from the immigration of Eurasian ancestors, likely coincident with Plio-Pleistocene climatic oscillations between arid and humid conditions.[84]
Phylogenetic tree of the extant wolf-like canids,[lower-alpha 1] with the pink shading representing the species Canis lupus.
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3.5 mya |
Admixture with an extinct unknown canid
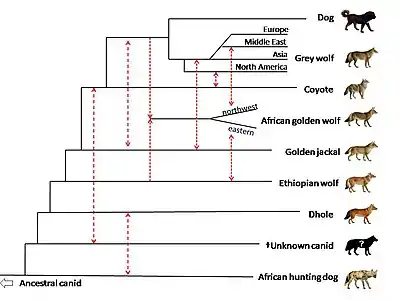
In 2018, whole genome sequencing was used to compare members of genus Canis, along with the dhole (Cuon alpinus) and the African hunting dog (Lycaon pictus). There is evidence of gene flow between African golden wolves, golden jackals, and gray wolves. The study suggests that the African golden wolf is a descendant of a genetically admixed canid of 72% grey wolf and 28% Ethiopian wolf ancestry, and that the Ethiopian wolf once had a wider range in Africa. One African golden wolf from the Egyptian Sinai Peninsula showed high admixture with the Middle Eastern gray wolves and dogs, highlighting the role of the land bridge between the African and Eurasian continents in canid evolution. There is evidence of gene flow between golden jackals and Middle Eastern wolves, less so with European and Asian wolves, and least with North American wolves. The study proposes that the golden jackal ancestry found in North American wolves may have occurred before the divergence of the Eurasian and North American gray wolves. The study indicates that the common ancestor of the coyote and gray wolf has genetically admixed with a ghost population of an extinct unidentified canid. The canid is genetically close to the dhole and has evolved after the divergence of the African hunting dog from the other canid species. The basal position of the coyote compared to the wolf is proposed to be due to the coyote retaining more of the mitochondrial genome of this unknown canid.[104]
A genomic study on the wolves of China included museum specimens of wolves from southern China that were collected between 1963 and 1988. The wolves in the study formed 3 clades: north Asian wolves that included those from northern China and eastern Russia, Himalayan wolves from the Tibetan Plateau, and a unique population from southern China. One specimen located as far southeast as Jiangxi province shows evidence of being admixed between Tibetan-related wolves and other wolves in China. One specimen from Zhejiang province in eastern China shared gene flow with the wolves from southern China, however its genome was 12-14 percent admixed with a canid that may be the dhole or an unknown canid that predates the genetic divergence of the dhole. The wolf population from southern China is believed to be still existing in that region.[118]
Two wolf haplogroups
A haplotype (haploid genotype) is a group of genes in an organism that are inherited together from a single parent.[119][120] A haplogroup is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism (a mutation).[121][122] Mitochondrial DNA passes along a maternal lineage that can date back thousands of years.[121]
In 2010, a study compared DNA sequences that were 230 base pairs in length from the mitochondrial control region of 24 ancient wolf specimens from western Europe dated between 44,000–1,200 YBP with those of modern gray wolves. Most of the sequences could be represented on a phylogenetic tree. However, the haplotypes of the Himalayan wolf and the Indian gray wolf could not because they were 8 mutations apart from the other wolves,[111] indicating distinct lineages which had previously been found in other studies.[111][108][109] The study found that there were 75 different gray wolf mDNA haplotypes that include 23 in Europe, 30 in Asia, 18 in North America, 3 in both Europe and Asia, and 1 in both Europe and North America.[111] These haplotypes could be allocated into two haplogroups[113][111] that were separated from each other by 5 mutations. Haplogroup 1 formed a monophyletic clade (indicating that they all carried the same mutation inherited from a single female ancestor). All other haplotypes were basal in the tree, and these formed 2–3 smaller clades that were assigned to haplogroup 2 that was not monophyletic.[111][123]
Haplogroups 1 and 2 could be found spread across Eurasia but only haplogroup 1 could be found in North America. The ancient wolf samples from western Europe differed from modern wolves by 1 to 10 mutations, and all belonged to haplogroup 2 indicating a haplogroup 2 predominance in this region for over 40,000 years before and after the Last Glacial Maximum. A comparison of current and past frequencies indicated that in Europe haplogroup 2 became outnumbered by haplogroup 1 over the past several thousand years[113] but in North America haplogroup 2 became extinct and was replaced by haplogroup 1 after the Last Glacial Maximum.[111][123] Access into North America was available between 20,000–11,000 years ago after the Wisconsin glaciation had retreated but before the Bering land bridge became inundated by the sea.[124] Therefore, haplogroup 1 was able to enter into North America during this period.
Stable isotope analysis conducted on the bone of a specimen allows researchers to form conclusions about the diet, and therefore the ecology, of extinct wolf populations. This analysis suggests that the Pleistocene wolves from haplogroup 2 found in Beringia and Belgium preyed mainly on Pleistocene megafauna,[64][111][110] which became rare at the beginning of the Holocene 12,000 years ago.[111][125] One study found the Beringian wolf to be basal to all other gray wolves except for the extant Indian gray wolf and the extant Himalayan wolf.[110] The Pleistocene Eurasian wolves have been found to be morphologically and genetically comparable to the Pleistocene eastern-Beringian wolves,[126] with some of the ancient European and Beringian wolves sharing a common haplotype (a17),[111][110] which makes ecological similarity likely.[111] Two ancient wolves from the Ukraine dated around 30,000 YBP and the 33,000 YBP "Altai dog" had the same sequence as six Beringian wolves, and another from the Czech Republic dated 44,000 YBP had the same sequence as two Beringian wolves.[110]
It has been proposed that the Pleistocene wolves across northern Eurasia and northern North America represented a continuous and almost panmictic population that was genetically and probably also ecologically distinct from the wolves living in this area today.[111][127] The specialized Pleistocene wolves did not contribute to the genetic diversity of modern wolves, and the modern wolf populations across the Holarctic are likely to be the descendants of wolves from populations that came from more southern refuges.[127] Extant haplogroup 2 wolves can be found in Italy, the Balkans and the Carpathian Mountains but rare elsewhere in Europe. In Asia, only four haplotypes have been identified as belonging to this haplogroup, and two of them occur in the Middle East.[128] Haplogroup 2 did not become extinct in Europe, and if before the Last Glacial Maximum haplogroup 2 was exclusively associated with the wolf ecomorph specialized in preying on megafauna, it would mean that in Europe it was capable of adapting to changing prey.[111]
In 2013, a mitochondrial DNA sequencing of ancient wolf-like canids revealed another separate lineage of 3 haplotypes (forming a haplogroup) that was found in 3 Late Pleistocene specimens from Belgium; however, it has not been detected in extant wolves.[66][128] One of these was the "Goyet dog".[66]
Dissenting view

In 2016, a study was undertaken due to concerns that previous mDNA studies may have been conducted with insufficient genetic resolution or limited geographical coverage and had not included sufficient specimens from Russia, China, and the Middle East. The study compared a 582 base pair sequence of the mitochondrial control region which gave twice the phylogenetic resolution of the 2010 study.[111] The study compared the sequences of both modern wolves and ancient wolf specimens, including specimens from the remote areas of North America, Russia and China. The study included the Taimyr wolves, the Goyet "dog", the Altai "dog", Beringian wolves, and other ancient specimens.[105]
The study found 114 different wolf haplotypes among 314 sequences, with the new haplotypes being found in Siberia and China. The phylogenetic tree resolved into 19 clades that included both modern and ancient wolves, which showed that the most basal clades included the Indian gray wolf and the Himalayan wolf, with a subclade of wolves from China and Mongolia falling within the Himalayan wolf clade. The two most basal North American haplotypes included the Mexican wolf and the Vancouver Island wolf, however the Vancouver Island wolf showed the same haplotype as a dog which indicates admixture,[105] with the dog lineage basal to all extant North American subspecies.[11] In Europe, the two most genetically distinct haplotypes form the Iberian wolf and separately the Italian wolf that was positioned close to the ancient wolves. The Greenland wolves all belonged to one haplotype that had been previously found among North American wolves and which indicates their origin from North America. The Eastern wolf was confirmed as a coyote/wolf hybrid. Wolves found in the regions of the Chukotka Peninsula, the North Korean border, Amur Oblast and Khakassia showed the greatest genetic diversity and with close links to all other wolves found across the holarctic. One ancient haplotype that had been found in Alaska (Eastern Beringia 28,000 YBP) and Russia (Medvezya "Bear" Cave, Pechora area, Northern Urals 18,000 YBP) was shared with some modern wolves found in China and Mongolia.[105]
The previous finding of two wolf haplogroups[111] was not clearly delineated in this study but it agreed that the genetic diversity of past wolves has been lost at the beginning of the Holocene in Alaska, Siberia, and Europe with limited overlap with modern wolves. For the ancient wolves of North America, instead of an extinction/replacement model suggested by a previous study,[110] this study found substantial evidence of a population bottleneck in North America in which the ancient wolf diversity was almost lost around the beginning of the Holocene (no further elaboration in the study). In Eurasia, the loss of ancient lineages could not be simply explained and appears to have been slow across time with the reasons unclear.[105]
Into America and Japan
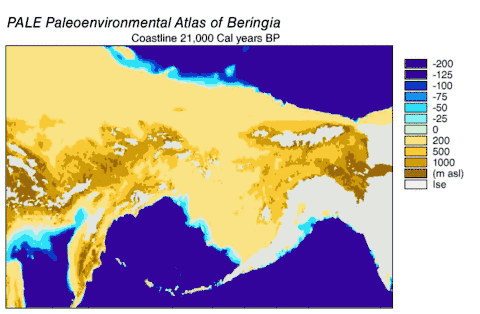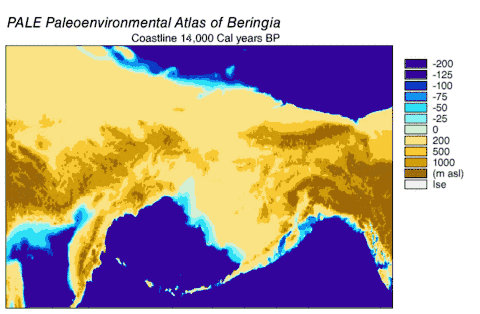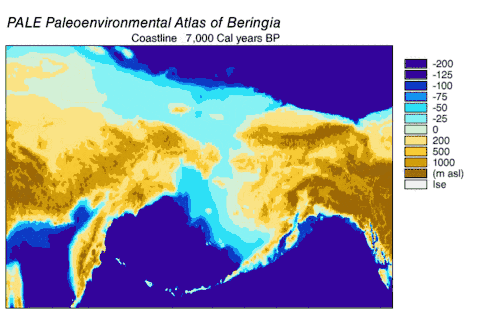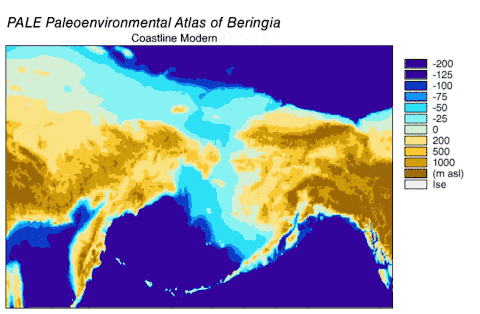
In 2016, a study built on the work of another major study[66] and analyzed the sequences of 12 genes that are located on the heavy strand of the mitochondrial genome of extinct and modern C. lupus. The study excluded the sequences of the divergent Himalayan wolf and the Indian gray wolf. The ancient specimens were radiocarbon dated and stratagraphically dated, and together with the sequences generated a time-based phylogenetic tree. From the tree, the study was able to infer the most recent common ancestor for all other C. lupus specimens – modern and extinct – was 80,000 YBP and this date concurred with the earlier study.[66][115] The study could find no evidence of a population bottleneck for wolves until a few thousand years ago.[115]
The phylogenetic tree showed the polyphyly of American wolves, the Mexican wolf was divergent from other North American wolves, and these other North American wolves formed two closely related clades. A scenario consistent with the phylogenetic, ice sheet and sea-level data was that during the Ice Age when sea levels were at their lowest, there was a single wave of wolf colonization into North America starting with the opening of the Bering land bridge 70,000 YBP and closing during the Late Glacial Maximum of the Yukon corridor that ran through the division between the Laurentide Ice Sheet and the Cordilleran Ice Sheet 23,000 YBP. Mexican wolves were part of the single wave and either diverged from the other wolves before entering North America or once in North America due to the change in its environment.
As wolves had been in the fossil record of North America but modern wolves could trace their ancestry back only 80,000 years, the wolf haplotypes that were already in North America were replaced by these invaders, either through competitive displacement or through admixture.[115] The replacement in North America of a basal population of wolves by a more recent one supported the findings of earlier studies.[111][123][110][115] There possibly existed a panmictic wolf population with gene flow spanning Eurasia and North America until the closing of the ice sheets.[111][127][115] Once the sheets closed, the southern wolves were isolated and north of the sheets only the Beringian wolf existed. The land bridge became inundated by the sea 10,000 YBP, the sheets receded 12,000–6,000 YBP, the Beringian wolf went extinct and the southern wolves expanded to recolonize the rest of North America. All North American wolves are descended from those that were once isolated south of the ice sheets. However, much of their diversity was later lost during the twentieth century.[115]
Studies using mitochondrial DNA have indicated that the wolves of coastal south-east Alaska are genetically distinct from inland gray wolves, reflecting a pattern also observed in other taxa. They show a phylogenetic relationship with extirpated wolves from the south (Oklahoma), indicating that these wolves are the last remains of a once widespread group that has been largely extirpated during the last century, and that the wolves of northern North America had originally expanded from southern refuges below the Wisconsin glaciation after the ice had melted at the end of the Last Glacial Maximum.[129][130][131] A whole-genome DNA study indicated that all North American wolves were monophyletic and therefore are the descendants of a common ancestor.[132]
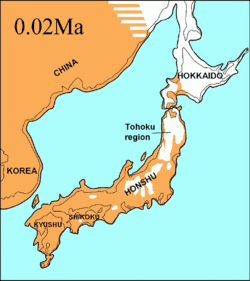
During the same period, the Soya Strait between Hokkaido and Sakhalin Island was dry for 75,000 years and it was proposed that the extinct Ezo wolf (C. l. hattai) arrived on Hokkaido from Sakhalin.[133][115][134] However, the sequences indicated that it arrived in Hokkaido less than 10,000 YBP. The Ezo wolf was closely related to one of the North American clades,[133][115][135] but different to the more southerly Japanese wolf (C. l. hodophilax) that was basal to modern wolves.[133][115] The Japanese wolf inhabited Kyushu, Shikoku, and Honshu islands[136][137] but not Hokkaido Island.[137] This indicates that its ancestor may have migrated from the Asian continent through the Korean Peninsula into Japan.[133][137] The past sea levels of the Korean Strait together with the timing of the Japanese wolf sequences indicated that it arrived to the southern islands less than 20,000 YBP.[115]
The dog was a very successful invader of North America and had established a widespread ecological niche by the Early–Middle Holocene. There was no overlap in niche between the dog and the wolf in comparison to the dog and other North American canids. By the Late Holocene, the dog's niche area was less in size than researchers had expected to find, indicating that it was limited by biotic factors. These regions include the northeast and northwest of the United States that correlate with the greatest densities of early human occupation, indicating that the dog had "defected" from the wolf niche to the human niche and explains why the dog's niche area was not as large as expected. The separation between dog and wolf may reflect the rapid rate in which domestication occurred,[138] including the possibility of a second domestication event occurring in North America.[139][138] Packs of wolves and hunter-gatherers hunt similar prey in a similar way within a similar group social structure that may have facilitated wolf domestication.[140][52]
The wolf was exterminated in the southern part of their historic geographical range in North America by the middle of the 20th century. An mDNA study of 34 wolf remains from North America dated between 1856 and 1915 found their genetic diversity to be twice that of modern wolves in these regions, and two thirds of the haplotypes identified were unique. These results indicate that a historic population of several hundred thousand wolves once existed in Mexico and the western US.[141][113]
Beringia
Grey wolves suffered a species-wide population bottleneck (reduction) approximately 25,000 YBP during the Last Glacial Maximum. This was followed by a single population of modern wolves expanding out of a Beringia refuge to repopulate the wolf's former range, replacing the remaining Late Pleistocene wolf populations across Eurasia and North America as they did so.[114][116][117] This source population probably did not give rise to dogs, but admixed with dogs which allowed them to gain coat colour genes that are also related to immunity, and provided dogs with genes which allowed them to adapt to high-altitude environments (e.g. Tibet). This suggests that the genetic divergence of European and East Asian dogs could be based on admixture with different sub-populations of wolves.[117]
There is little genetic information available on the ancient wolves that existed prior to the bottleneck. However, studies show that one or more of these ancient populations is more directly ancestral to dogs than are modern wolves, and conceivably these were more prone to domestication by the first humans to invade Eurasia.[117]
As of 2020, the oldest known intact wolf remains belongs to a mummified pup dated 56,000 YBP that was recovered from the permafrost along a small tributary of Last Chance Creek near Dawson City, Yukon, Canada. A DNA analysis showed that it belonged to the Beringian wolf clade, that the most recent common ancestor of this clade dates to 86,700–67,500 YBP, and that this clade was basal to all other wolves except for the Himalayan wolf.[142]
Divergence with the coyote
In 1993, a study proposed that the wolves of North America display skull traits more similar towards the coyote than those wolves from Eurasia.[41] In 2016, a whole-genome DNA study proposed, based on the assumptions made, that all of the North American wolves and coyotes diverged from a common ancestor less than 6,000–117,000 years ago. The study also indicated that all North America wolves have a significant amount of coyote ancestry and all coyotes some degree of wolf ancestry, and that the red wolf and eastern wolf are highly admixed with different proportions of gray wolf and coyote ancestry. One test indicated a wolf/coyote divergence time of 51,000 years before present that matched other studies indicating that the extant wolf came into being around this time. Another test indicated that the red wolf diverged from the coyote between 55,000–117,000 years before present and the Great Lakes region wolf 32,000 years before present. Other tests and modelling showed various divergence ranges and the conclusion was a range of less than 6,000 and 117,000 years before present.[132][143] This finding conflicts with the fossil record that indicates a coyote-like specimen dated to 1 million years before present.[15]
The modern gray wolf expanded out of Beringia 25,000 years ago. This implies that if the coyote and red wolf were derived from this invasion, their histories date only tens of thousands and not hundreds of thousands of years ago, which is consistent with other studies.[117]
Domestic dog
The domestic dog (Canis lupus familiaris) is the most widely abundant large carnivore.[66][11][144] Over the past million years, numerous wolf-like forms existed but their turnover has been high, and modern wolves are not the lineal ancestors of dogs.[66][9][11][141] Although research had suggested that dogs and wolves were genetically very close relatives,[85][86][95] later phylogenetic analysis strongly supported the hypothesis that dogs and wolves are reciprocally monophylic taxa that form two sister clades.[85][9][145] This suggests that none of the modern wolf populations are related to the wolves that were first domesticated and the wolf ancestor of dogs is therefore presumed extinct.[9][145] Recent mitochondrial DNA analyses of ancient and modern gray wolf specimens supports a pattern of population reduction and turnover.[66][111][110] An alternate proposal is that during the ecological upheavals of the Late Pleistocene all of the remaining members of a dwindling lineage joined humans.[146][67]
In 2016, a study investigated for the first time the population subdivisions, demography, and the relationships of gray wolves based on their whole-genome sequences. The study indicated that the dog was a divergent subspecies of the gray wolf and was derived from a now-extinct ghost population of Late Pleistocene wolves,[66][9][11] and the dog and the dingo are not separate species.[11] The genome-wide phylogenetic tree indicated a genetic divergence between New World and Old World wolves, which was then followed by a divergence between the dog and Old World wolves 27,000YBP[10][11] – 29,000 YBP.[11] The dog forms a sister taxon with Eurasian gray wolves but not North American wolves. The dog had considerable pre-ancestry after its divergence from the Old World wolves before it separated into distinct lineages that are nearly as distinct from one another as they are from wolves.[11] The study suggested that previous datings based on the divergence between wolves and coyotes of one million years ago using fossils of what appeared to be coyote-like specimens may not reflect the ancestry of the modern forms.[84][9][10][11]
| Gray wolf divergence and timing | ||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Whole-genome phylogenetic tree – extant gray wolf populations,[11] with divergence times calculated using an assumed mutation rate of Lindblad-Toh (1x10−8)[83] or [Skoglund] (0.4x10−8).[10] The dog is a genetically divergent subspecies of the gray wolf, with substantial divergence between the dog lineages as nearly as distinct from one another as wolves are from dogs, which may reflect more admixture and wolf ancestry retained in their genome.[11] |
The study indicated that the Mexican wolf was also a divergent form of gray wolf, suggesting that may have been part of an early invasion into North America.[11][141] The Tibetan wolf was found to be the most highly divergent of the Old World wolves, had suffered a historical population bottleneck and had only recently recolonized the Tibetan Plateau. Glaciation may have caused its habitat loss, genetic isolation then local adaption.[11]
The study indicated that there has been extensive genetic admixture between domestic dogs and wolves, with up to 25% of the genome of Old World wolves showing signs of dog ancestry, possibly as the result of gene flow from dogs into wolves that were ancestral to all modern wolves. There was evidence of significant gene flow between the European wolves plus the Israeli wolf with the basenji and boxer, which suggests admixture between the lineages ancestral to these breeds and wolf populations.[9][11] For the lowland Asian wolves: the Central Russian and East Russian wolves and all of the lowland Chinese wolves had significant gene flow with the Chinese indigenous dogs, the Tibetan Mastiff and the dingo. For the highland Asian wolves: The Tibetan wolves did not show significant admixture with dogs; however, the Qinghai wolves had gene flow with the dingo and one of them had gene flow with the Chinese dogs. The New World wolves did not show any gene flow with the boxer, dingo or Chinese indigenous dogs but there was indication of gene flow between the Mexican wolf and the African basenji.[11] All species within the genus Canis, the wolf-like canids, are phylogenetically closely related with 78 chromosomes and can potentially interbreed.[86] There was indication of gene flow into the golden jackal from the population ancestral to all wolves and dogs (11.3%–13.6%) and much lower rates (up to 2.8%) from extant wolf populations.[9][11]
The data indicated that all wolves shared similar population trajectories, followed by population decline that coincided with the expansion of modern humans worldwide and their technology for capturing large game.[11][147] Late Pleistocene carnivores would have been social living in large prides, clans and packs in order to hunt the larger game available at that time, and these larger groups would have been more conspicuous targets for human persecutors.[147] Large dogs accompanying the humans may have accelerated the rate of decline of carnivores that competed for game,[11][148] therefore humans expanded across Eurasia, encountered wolves, domesticated some and possibly caused the decline of others.[11]
The study concluded that admixture had confounded the ability to make inferences about the place of dog domestication. Past studies based on SNPs, genome-wide similarities with Chinese wolves, and lower linkage disequilibrium might reflect regional admixture between dogs with wolves and gene flow between dog populations, with divergent dog breeds possibly maintaining more wolf ancestry in their genome. The study proposed that analysis of ancient DNA might be a better approach.[11]
In the same year, a study found that there were only 11 fixed genes that showed variation between wolves and dogs. These genes are thought to affect tameness and emotional processing ability.[149] Another study provided a listing of all of the gray wolf and dog mDNA haplotypes combined in the one phylogenetic tree.[150]
In 2018, a study compared the sequences of 61,000 Single-nucleotide polymorphisms (mutations) taken from across the genome of grey wolves. The study indicated that there exists individual wolves of dog/wolf ancestry in most of the wolf populations of Eurasia but less so in North America. The hybridization has been occurring across different time scales and was not a recent event. Low-level hybridization did not reduce the wolf distinctiveness.[151]
Dingo
The dingo (Canis lupus dingo) refers to the dog found in Australia. The dingo is a divergent subspecies of the gray wolf and is not a separate species,[11] and is considered genetically to be a basal member of the domestic dog clade.[11][84][9] The genetic evidence indicates that the dingo originated from East Asian domestic dogs and was introduced through the South-East Asian archipelago into Australia,[153][154] with a common ancestry between the Australian dingo and the New Guinea Singing Dog.[154][155]
Taimyr wolf

In May 2015 a study was conducted on a partial rib-bone of a wolf specimen (named "Taimyr-1") found near the Bolshaya Balakhnaya River in the Taimyr Peninsula of Arctic North Asia, that was AMS radiocarbon dated to 34,900 YBP. The sample provided the first draft genome of the cell nucleus for a Pleistocene carnivore, and the sequence was identified as belonging to Canis lupus.[10]
Using the Taimyr-1 specimen's radiocarbon date, its genome sequence and that of a modern wolf, a direct estimate of the genome-wide mutation rate in dogs / wolves could be made to calculate the time of divergence. The data indicated that the previously unknown Taimyr-1 lineage was a wolf population separate to modern wolves and dogs and indicated that the Taimyr-1 genotype, gray wolves and dogs diverged from a now-extinct common ancestor[10][73][156] before the peak of the Last Glacial Maximum, 27,000–40,000 years ago. The separation of the dog and wolf did not have to coincide with selective breeding by humans.[10][157] Such an early divergence is consistent with several paleontological reports of dog-like canids dated up to 36,000 YBP, as well as evidence that domesticated dogs most likely accompanied early colonizers into the Americas.[10]
Comparison to the gray wolf lineage indicated that Taimyr-1 was basal to gray wolves from the Middle East, China, Europe and North America but shared a substantial amount of history with the present-day gray wolves after their divergence from the coyote. This implies that the ancestry of the majority of gray wolf populations today stems from an ancestral population that lived less than 35,000 years ago but before the inundation of the Bering Land Bridge with the subsequent isolation of Eurasian and North American wolves.[10]
A comparison of the ancestry of the Taimyr-1 lineage to the dog lineage indicated that some modern dog breeds have a closer association with either the gray wolf or Taimyr-1 due to admixture. The Saarloos wolfdog showed more association with the gray wolf, which is in agreement with the documented historical crossbreeding with gray wolves in this breed. Taimyr-1 shared more alleles (gene expressions) with those breeds that are associated with high latitudes: the Siberian husky and Greenland dog[10][156] that are also associated with arctic human populations, and to a lesser extent the Shar Pei and Finnish spitz. An admixture graph of the Greenland dog indicates a best-fit of 3.5% shared material, although an ancestry proportion ranging between 1.4% and 27.3% is consistent with the data. This indicates admixture between the Taimyr-1 population and the ancestral dog population of these four high-latitude breeds. These results can be explained either by a very early presence of dogs in northern Eurasia or by the genetic legacy of Taimyr-1 being preserved in northern wolf populations until the arrival of dogs at high latitudes. This introgression could have provided early dogs living in high latitudes with phenotypic variation beneficial for adaption to a new and challenging environment. It also indicates that the ancestry of present-day dog breeds descends from more than one region.[10]
An attempt to explore admixture between Taimyr-1 and gray wolves produced unreliable results.[10]
As the Taimyr wolf had contributed to the genetic makeup of the Arctic breeds, a later study suggested that descendants of the Taimyr wolf survived until dogs were domesticated in Europe and arrived at high latitudes where they mixed with local wolves, and these both contributed to the modern Arctic breeds. Based on the most widely accepted oldest zooarchaeological dog remains, domestic dogs most likely arrived at high latitudes within the last 15,000 years. The mutation rates calibrated from both the Taimyr wolf and the Newgrange dog genomes suggest that modern wolf and dog populations diverged from a common ancestor between 20,000–60,000 YBP. This indicates that either dogs were domesticated much earlier than their first appearance in the archaeological record, or they arrived in the Arctic early, or both.[158]
The finding of a second wolf specimen from the same area (“Taimry-2”) and dated to 42,000 YBP has also been sequenced but yielded only mitochondrial DNA.[159]
Canis variabilis
In 2015, a study looked at the mitochondrial control region sequences of 13 ancient canid remains and one modern wolf from five sites across Arctic north-east Siberia. The fourteen canids revealed nine mitochondrial haplotypes, three of which were on record and the others not reported before. The phylogentic tree generated from the sequences showed that four of the Siberian canids dated 28,000 YBP and one Canis c.f. variabilis dated 360,000 YBP were highly divergent. The haplotype designated as S805 (28,000 YBP) from the Yana River was one mutation away from another haplotype S902 (8,000 YBP) that represents Clade A of the modern wolf and domestic dog lineages. Closely related to this haplotype was one that was found in the recently extinct Japanese wolf. Several ancient haplotypes were oriented around S805, including Canis c.f. variabilis (360,000 YBP), Belgium (36,000 YBP – the "Goyet dog"), Belgium (30,000 YBP), and Konsteki, Russia (22,000 YBP). Given the position of the S805 haplotype on the phylogenetic tree, it may potentially represent a direct link from the progenitor (including Canis c.f. variabilis) to the domestic dog and modern wolf lineages. The gray wolf is thought to be ancestral to the domestic dog, however its relationship to C. variabilis, and the genetic contribution of C. variabilis to the dog, is the subject of debate.[160]
The Zhokhov Island (8,700 YBP) and Aachim (1,700 YBP) canid haplotypes fell within the domestic dog clade, cluster with S805, and also share their haplotypes with – or are one mutation away from – the Tibetan wolf (C. l. filchneri) and the recently extinct Japanese wolf (C. l. hodophilax). This may indicate that these canids retained the genetic signature of admixture with regional wolf populations. Another haplotype designated as S504 (47,000 YBP) from Duvanny Yar appeared on the phylogenetic tree as not being connected to wolves (both ancient and modern) yet ancestral to dogs, and may represent a genetic source for regional dogs.[160]
The authors concluded that the structure of the modern dog gene pool was contributed to from ancient Siberian wolves and possibly from Canis c.f. variabilis.[160][161]
Rise to dominant predator
In 2015, a study looked at the paleoecology of large carnivores across the Mammoth steppe during the Late Pleistocene by using stable isotope analysis of their fossil collagen to reconstruct their diets. Based on testing in Belgium, around 40,000 YBP the Cave hyenas preyed on mammoth, woolly rhinoceros, horses and reindeer, with cave lions taking reindeer and young cave bears. Wolves appear to have been out-competed by cave hyenas and had their diet restricted to chamois, giant deer and red deer. However, after the Last Glacial Maximum around 14,000 YBP, wolves had access to all prey species, the cave lion was restricted to reindeer, and the cave hyena had gone extinct.[162][163][164] The data suggests that the extinction of the cave hyena allowed the wolf to become the dominant predator rather than the cave lion, just before the cave lion's extinction.[164] Another study indicated that the wolf thrived compared to the cave hyena when there was greater snow cover.[165]
Wolf population differences
The grey wolf Canis lupus is a highly adaptable species that is able to exist in a range of environments and which possesses a wide distribution across the Holarctic. Studies of modern grey wolves have identified distinct sub-populations that live in close proximity to each other.[2][3] This variation in sub-populations is closely linked to differences in habitat – precipitation, temperature, vegetation, and prey specialization – which affect cranio-dental plasticity.[4][5][6][7] The archaeological and paleontological records show their continuous presence for at least the last 300,000 years.[8] This continuous wolf presence contrasts with genomic studies, which suggest that all modern wolves and dogs descend from a common ancestral wolf population[9][10][11] that existed as recently as 20,000 years ago.[9] These studies indicate that a population bottleneck was followed by a rapid radiation from an ancestral population at a time during, or just after, the Last Glacial Maximum. This implies that the original wolf populations were out-competed by a new type of wolf which replaced them.[115] However, the geographic origin of this radiation is not known.
Apart from domestication, humans have harmed the wolf by restricting its habitat through persecution. This has caused a dramatic decrease in its population size over the last two centuries.[166][167] The shrinking of its habitats that overlap with those of close-relatives such as dogs and coyotes have led to numerous occurrences of hybridization.[168][169] These events, in addition to recent turnovers (extinctions and repopulations by other geneotypes), has made the unravelling of the phylogeographic history of the wolf difficult.[105]
Ecotypes
An ecotype is a variant in which the phenotypic differences are too few or too subtle to warrant being classified as a subspecies. These can occur in the same geographic region where distinct habitats such as meadow, forest, swamp, and sand dunes provide ecological niches. Where similar ecological conditions occur in widely separated places it is possible for a similar ecotype to occur. This is different to a subspecies, which may exist across a number of different habitats. In animals, ecotypes can be regarded as micro-subspecies that owe their differing characteristics to the effects of a very local environment.[170] Ecotypes have no taxonomic rank.
Gray wolves have a wide, natural distribution across the Holarctic that includes many different habitats, which can vary from the high arctic to dense forests, open steppe and deserts. The genetic differences between different populations of gray wolves is tightly linked to the type of habitat in which they live.[7] Differences in genetic markers among the Scandinavian wolf population has arisen in only just over a decade due to their small population size,[7][171] which indicates that these differences are not dependent on a long time spent in isolation and that larger population patterns can evolve in just a few thousand years.[7] These differences can also include fur color and density, and body size.[7][172][2] The differences can also include behavior, as coastal wolves eat fish[7][172] and tundra wolves migrate.[7][2] These differences have been observed between two wolf populations that are living in close proximity. It has been shown that mountain wolves do not interbreed with nearby coastal wolves, and the Alps of France and Switzerland have been repopulated with wolves from the mountains of nearby Italy[7][173] and from the far away mountains of Croatia[7][174] rather than from the nearer lowlands, which indicates that distance is not the driving force in differences between the two ecomorphs.[7]
In 2013, a genetic study found that the wolf population in Europe was divided along a north–south axis and formed five major clusters. Three clusters were identified occupying southern and central Europe in Italy, the Carpathians, and the Dinaric-Balkans. Another two clusters were identified occupying north-central Europe and the Ukrainian steppe. The Italian wolf consisted of an isolated population with low genetic diversity. Wolves from Croatia, Bulgaria, and Greece formed the Dinaric-Balkans cluster. Wolves from Finland, Latvia, Belarus, Poland and Russia formed the north-central Europe cluster with wolves from the Carpathians cluster a mixture of wolves from the north-central cluster and the Dinaric-Balkans cluster. The wolves from the Carpathians were more similar to the wolves from the Ukrainian Steppe than they were to wolves from north-central Europe. These clusters may have been the result of expansion from glacial refugia, an adaptation to local environments, and landscape fragmentation and the killing of wolves in some areas by humans.[175]
In 2016, two studies compared the sequences of 42,000 single nucleotide polymorphisms in North American gray wolves and found that they formed six ecotypes. These six wolf ecotypes were named West Forest, Boreal Forest, Arctic, High Arctic, Baffin, and British Columbia. The studies found that precipitation and mean diurnal temperature range were the most influential variables on sequence variation.[3][176] These findings were in accord with previous findings that precipitation influenced morphology,[177] and that vegetation[4] and habitat type[172][178] influenced wolf differences. One of these studies found that the variation in 11 key genes affected wolf vision, sense of smell, hearing, coat color, metabolism, and immunity. The study identified 1,040 genes that are potentially under selection due to habitat variation, and therefore that there was evidence of local adaption of the wolf ecotypes at a molecular level. Most notable was the positive selection of genes that influence vision, coat color, metabolism and immunity in the Arctic and High Arctic ecotypes, and that the British Columbia ecotype also has a unique set of adaptions.[176] The local adaptation of a wolf ecotype most likely reflects a wolf's preference to remain in the type of habitat that it was born into.[3]
Ecological factors including habitat type, climate, prey specialization and predatory competition will greatly influence gray wolf genetic population structure and cranio-dental plasticity.[125][73][7][2][179][180][4][5][6] During the Last Glacial Maximum, there was greater wolf genetic diversity than there is today,[9][66] and within the Pleistocene gray wolf population the variations between local environments would have encouraged a range of wolf ecotypes that were genetically, morphologically and ecologically distinct from one another.[73]
Pleistocene wolves
The oldest Canis remains found in Europe were from France and dated to 3.1 million YBP,[181] followed by Canis cf. etruscus (where cf. in Latin means confer, uncertain) from Italy dated to 2.2 million YBP.[182] C. lupus first appeared in Italy during Marine Isotope Stage 9[183] (337,000 YBP). In Britain, it was the only canid species present from MIS 7 (243,000 YBP), with the oldest record from Pontnewydd Cave in north Wales.[184] During the Ice Age, Britain was separated from Europe by only the Channel River.
A study of Pleistocene C. lupus in Britain at different time periods found that its abilities to crush, slice meat and eat bone highlighted its cranio-dental plasticity. These responses to dietary changes showed species-wide dietary shifts, and not just local ecomorphs, in response to climatic and ecological variables. The survival of C. lupus during the Pleistocene can be attributed largely to its plastic cranio-dental morphology.[6]
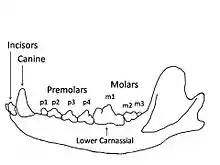

| Time YBP | Variables |
|---|---|
| 243,000 MIS 7 | Paleoenvironment was open grasslands with summer temperatures between 16 °C and 23 °C and winter temperatures between −7 °C and −6 °C dominated by steppe mammoth and horse. Competitors included the lion, brown bear, and rarely the spotted hyena. The wolves of MIS 7 were slightly smaller in body size than MIS 5 wolves and those found in Sweden today. These wolves were out-competed by the larger competitors, leading to a more omnivorous diet with increased crushing ability in an open environment that supported more types of prey and more non-meat foods than the MIS 5 period. They had shallower and narrower jaws than MIS 5 wolves and those found in Sweden today, which indicated that they could take only small to medium-sized prey. They exhibited a lower percentage of tooth breakage comparable with MIS-3 wolves. However, they had the highest percentage of moderately worn teeth.[6] |
| 82,000 MIS 5A | Paleoenvironment was cold, open tundra with summer temperatures between 7 °C and 11 °C and winter temperatures between −10 °C and −30 °C dominated by reindeer and bison. A large form of brown bear was top predator, with no hyena at this time. The wolves of MIS 5 were larger in body size than those found in Sweden today. These wolves suffered from a severe climate, low prey availability and dietary stress leading to a more carnivorous diet, with increased scavenging of frozen carcasses and bone consumption. They developed strong jaws and the highest flesh-slicing ability compared to the other wolves, with shallower jaws than the modern wolf but broader and deeper jaws than MIS 7 and MIS 5 wolves. They exhibited the longest and narrowest upper P4 that suggests improved slicing ability, and longest upper M1 and M2 but with reduced width and therefore reduced crushing ability, indicating a hypercarnivore. They exhibited a higher percentage of tooth breakage and severely worn teeth compared to the other wolves, and may have been using their upper P4 and lower m1 to crush bone rather than their molars, leading to a higher frequency of damage.[6] |
| 57,000 MIS 3 | Paleoenvironment of open grasslands with summer temperature of around 12 °C and winter temperature around −20 °C dominated by woolly mammoth, woolly rhinoceros, horse, and giant deer. Competitors included the lion, brown bear, and the spotted hyena as the top carnivore. The wolves of MIS 3 were smaller in body size than MIS 5 wolves and those found in Sweden today. These wolves were out-competed by the lion and hyena, leading to a more omnivorous diet with increased crushing ability in an open environment that supported more types of prey and more non-meat foods than the MIS 5 period. They had shallower and narrower jaws than MIS 5 wolves and those found in Sweden today, which indicated that they could take only small to medium-sized prey. They exhibited a lower percentage of tooth breakage comparable with MIS-7 wolves with moderate tooth wear.[6] |
| Today (Sweden) | Wolves have been extirpated in Britain but not in Sweden, where the temperatures are similar to those of Britain during the MIS 7 period. Environment of boreal forest with summer temperatures between 14 °C and 18 °C and winter temperatures between 1 °C and −10 °C. The prey species includes elk, reindeer, roe deer, boar, hares, rabbit, and beaver. Competitors include the brown bear and lynx but the wolf is top carnivore. The wolves found in Sweden today are smaller in body size than MIS 5 wolves but larger than those of MIS 7 and MIS 3. The upper M1 and M2 length is longer than for MIS 7 and MIS 3 wolves, and the jaws deeper and broader, which indicates the ability to hunt and subdue large prey. However, the large molars retained a crushing ability and to process non-meat foods. These wolves live in boreal forests where small to medium game is hard to detect and labour-intensive to subdue, leading to an adaption for hunting larger game with higher reward. They are hypercarnivores similar to MIS 5 wolves but not with the same slicing ability.[6] |
During the Last Glacial Maximum 20,000 YBP, the Pleistocene steppe stretched across northern and central Eurasia and through Beringia into North America. The Pleistocene wolves of Beringia, and perhaps those across the steppe, were adapted to this habitat. Their tooth and skull morphology indicates that they specialized in preying on now-extinct Pleistocene megafauna, and their tooth wear indicates that their behavior was different to modern wolves.[110][7][185][186] This highlights the success of C. lupus as a species in adapting to different environmental conditions.[6] This gray wolf ecomorph became extinct at the end of the glaciation, along with the horse and other species on which it depended, and was replaced by wolves from southern North America. This indicates that specialized wolf ecomorphs can become extinct when their environment changes even though the habitat may still support other wolves.[7] Wolves went through a population bottleneck 20,000 YBP that coincides with the Last Glacial Maximum,[9][128][73][7] which indicates that many wolf populations may have gone extinct at the same time as the Beringian wolves.[7]
There are a small number of Canis remains that have been found at Goyet Cave, Belgium (36,500 YBP)[64] Razboinichya Cave, Russia (33,500 YBP)[187] Kostenki 8, Russia (33,500–26,500 YBP)[188] Predmosti, Czech Republic (31,000 YBP)[189] and Eliseevichi 1, Russia (17,000 YBP).[65] Based on cranial morphometric study of the characteristics thought to be associated with the domestication process, these have been proposed as early Paleolithic dogs.[188] These characteristics of shortened rostrum, tooth crowding, and absence or rotation of premolars have been documented in both ancient and modern wolves.[110][73][6][190][191][192] Rather than representing early dogs, these specimens may represent "a morphologically distinct local, now extinct, population of wolves".[73][193]
Notes
- Phylogenetic relationships and time of genetic divergence for the extant wolf-like canids based on DNA of the entire cell nucleus.[104] Exception 1: Side-striped jackal and Black-backed jackal added based on segments of DNA taken from the cell nucleus[83][84] with proposed divergence times in millions of years.[84] Exception 2: The position of the Himalayan wolf is based on segments of mitochondrial DNA[84][105][106][107] and segments from the cell nucleus.[84][106][107] Exception 3: The position of the Indian plains wolf is based on mDNA.[108][109][110][111][112][113] and its timing 120,000 years ago.[114] Exception 4: Late Pleistocene wolf included for time-line purposes only - the most recent common ancestor for all other Canis lupus specimens – modern and extinct but excluding the Indian plains wolf and the Himalayan wolf – was 80,000 years before present.[66][115] Exception 5: Gray wolf and dog timing 25,000 years ago.[114][116][117]
References
- Dawkins, William Boyd, Sir; Sanford, W. Ayshford; Reynolds, Sidney H. (1912). "British Pleistocene Hyænidæ, Ursidæ, Canidæ, and Mustelidæ". A Monograph of the British Pleistocene Mammalia. 2. London: Palaeontographical Society.
- Musiani, Marco; Leonard, Jennifer A.; Cluff, H. Dean; Gates, C. Cormack; Mariani, Stefano; Paquet, Paul C.; Vilà, Carles; Wayne, Robert K. (2007). "Differentiation of tundra/taiga and boreal coniferous forest wolves: Genetics, coat colour and association with migratory caribou". Molecular Ecology. 16 (19): 4149–70. doi:10.1111/j.1365-294X.2007.03458.x. PMID 17725575. S2CID 14459019.
- Schweizer, Rena M.; Vonholdt, Bridgett M.; Harrigan, Ryan; Knowles, James C.; Musiani, Marco; Coltman, David; Novembre, John; Wayne, Robert K. (2016). "Genetic subdivision and candidate genes under selection in North American grey wolves". Molecular Ecology. 25 (1): 380–402. doi:10.1111/mec.13364. PMID 26333947. S2CID 7808556.
- Geffen, ELI; Anderson, Marti J.; Wayne, Robert K. (2004). "Climate and habitat barriers to dispersal in the highly mobile grey wolf". Molecular Ecology. 13 (8): 2481–90. doi:10.1111/j.1365-294X.2004.02244.x. PMID 15245420. S2CID 4840903.
- Pilot, Malgorzata; Jedrzejewski, Wlodzimierz; Branicki, Wojciech; Sidorovich, Vadim E.; Jedrzejewska, Bogumila; Stachura, Krystyna; Funk, Stephan M. (2006). "Ecological factors influence population genetic structure of European grey wolves". Molecular Ecology. 15 (14): 4533–53. doi:10.1111/j.1365-294X.2006.03110.x. PMID 17107481. S2CID 11864260.
- Flower, Lucy O.H.; Schreve, Danielle C. (2014). "An investigation of palaeodietary variability in European Pleistocene canids". Quaternary Science Reviews. 96: 188–203. Bibcode:2014QSRv...96..188F. doi:10.1016/j.quascirev.2014.04.015.
- Leonard, Jennifer (2014). "Ecology drives evolution in grey wolves" (PDF). Evolution Ecology Research. 16: 461–473.
- Sotnikova, M (2010). "Dispersal of the Canini (Mammalia, Canidae: Caninae) across Eurasia during the Late Miocene to Early Pleistocene". Quaternary International. 212 (2): 86–97. Bibcode:2010QuInt.212...86S. doi:10.1016/j.quaint.2009.06.008.
- Freedman, Adam H.; Gronau, Ilan; Schweizer, Rena M.; Ortega-Del Vecchyo, Diego; Han, Eunjung; Silva, Pedro M.; Galaverni, Marco; Fan, Zhenxin; Marx, Peter; Lorente-Galdos, Belen; Beale, Holly; Ramirez, Oscar; Hormozdiari, Farhad; Alkan, Can; Vilà, Carles; Squire, Kevin; Geffen, Eli; Kusak, Josip; Boyko, Adam R.; Parker, Heidi G.; Lee, Clarence; Tadigotla, Vasisht; Siepel, Adam; Bustamante, Carlos D.; Harkins, Timothy T.; Nelson, Stanley F.; Ostrander, Elaine A.; Marques-Bonet, Tomas; Wayne, Robert K.; et al. (2014). "Genome Sequencing Highlights the Dynamic Early History of Dogs". PLOS Genetics. 10 (1): e1004016. doi:10.1371/journal.pgen.1004016. PMC 3894170. PMID 24453982.
- Skoglund, Pontus; Ersmark, Erik; Palkopoulou, Eleftheria; Dalén, Love (2015). "Ancient Wolf Genome Reveals an Early Divergence of Domestic Dog Ancestors and Admixture into High-Latitude Breeds". Current Biology. 25 (11): 1515–1519. doi:10.1016/j.cub.2015.04.019. PMID 26004765.
- Fan, Zhenxin; Silva, Pedro; Gronau, Ilan; Wang, Shuoguo; Armero, Aitor Serres; Schweizer, Rena M.; Ramirez, Oscar; Pollinger, John; Galaverni, Marco; Ortega Del-Vecchyo, Diego; Du, Lianming; Zhang, Wenping; Zhang, Zhihe; Xing, Jinchuan; Vilà, Carles; Marques-Bonet, Tomas; Godinho, Raquel; Yue, Bisong; Wayne, Robert K. (2016). "Worldwide patterns of genomic variation and admixture in gray wolves". Genome Research. 26 (2): 163–73. doi:10.1101/gr.197517.115. PMC 4728369. PMID 26680994.
- Sardella, Raffaele; Bertè, Davide; Iurino, Dawid Adam; Cherin, Marco; Tagliacozzo, Antonio (2014). "The wolf from Grotta Romanelli (Apulia, Italy) and its implications in the evolutionary history of Canis lupus in the Late Pleistocene of Southern Italy". Quaternary International. 328–329: 179–195. Bibcode:2014QuInt.328..179S. doi:10.1016/j.quaint.2013.11.016.
- Flynn, John J.; Wesley-Hunt, Gina D. (2005). "Phylogeny of the Carnivora: Basal Relationships Among the Carnivoramorphans, and Assessment of the Position of 'Miacoidea' Relative to Carnivora". Journal of Systematic Palaeontology. 3: 1–28. doi:10.1017/s1477201904001518. S2CID 86755875.
- "Eucyon davisi". Fossilworks. Retrieved 11 July 2016.
- Wang, Xiaoming; Tedford, Richard H.; Dogs: Their Fossil Relatives and Evolutionary History. New York: Columbia University Press, 2008.
- Vislobokova, I., Sotnikova, M. & Dodonov, A., 2003 – Bio-events and diversity of the Late Miocene-Pliocene mammal faunas of Russia and adjacent areas – in: Reumer, J.W.F. & Wessels, W. (eds.) – DISTRIBUTION AND MIGRATION OF TERTIARY MAMMALS IN EURASIA. A VOLUME IN HONOUR OF HANS DE BRUIJN – DEINSEA 10: 563–574 [ISSN 0923-9308] Published 1 December 2003
- R.M. Nowak (2003). "Chapter 9 - Wolf evolution and taxonomy". In Mech, L. David; Boitani, Luigi (eds.). Wolves: Behaviour, Ecology and Conservation. University of Chicago Press. pp. 239–258. ISBN 978-0-226-51696-7.
- Nowak, R. M. (1979). North American Quaternary Canis. 6. Monograph of the Museum of Natural History, University of Kansas. pp. 1–154. ISBN 978-0-89338-007-6.
- Wayne, R. K. (1995). "Red wolf: To conserve or not to conserve" (PDF). In Macdonald, D.; Handoca, L. (eds.). Canid News. 3. Newsletter of the IUCN/SSC Specialist Group. pp. 7–12.
- Hoffmeister, D. F., and W. W. Goodpaster. 1954. The mammals of the Huachuca Mountains, southeastern Arizona. Illinois Biological Monographs 24:1— 152.
- Lawrence, B.; Bossert, W. H. (1967). "Multiple character analysis of Canis lupus, latrans and niger". American Zoologist. 7 (2): 223–232. doi:10.1093/icb/7.2.223. JSTOR 3881428.
- Lawrence, B.; Bossert, W. H. (1975). "Relationships of North American Canis shown by a multiple character analysis of selected populations". In Fox, M. W. (ed.). Wild Canids: Their Systematics, Behavioral Ecology & Evolution. New York: Van Nostrand-Reinhold. ISBN 978-0-442-22430-1.
- Martin, R. A.; Webb, S. D. (1974). "Late Pleistocene mammals from the Devil's Den fauna, Levy County". In Webb, S. D. (ed.). Pleistocene Mammals of Florida. Gainesville: University Presses of Florida. pp. 114–145. ISBN 978-0-8130-0361-0.
- Webb, S. D. (1974). "Chronology of Florida Pleistocene mammals". Pleistocene Mammals of Florida. Gainesville: University Presses of Florida. pp. 5–31. ISBN 978-0-8130-0361-0.
- Wayne, R. K.; Jenks, S. M. (1991). "Mitochondrial DNA analysis supports extensive hybridization of the endangered red wolf (Canis rufus)". Nature. 351 (6327): 565–68. Bibcode:1991Natur.351..565W. doi:10.1038/351565a0. S2CID 4364642.
- "Canis lepophagus". Fossilworks. Retrieved 11 July 2016.
- Kurten, B. (1974). "A History of Coyote-Like Dogs (Canidae, Mamalia)". Acta Zoologica Fennica (140): 1–38.
- Kurten, B.; Anderson, E. (1980). Pleistocene mammals of North America. New York: Columbia University Press. pp. 1–442. ISBN 978-0-231-03733-4.
- Johnston, C. S. (1938). "Preliminary report on the vertebrate type locality of Cita Canyon and the description of an ancestral coyote". American Journal of Science. 5. 35 (209): 383–390. Bibcode:1938AmJS...35..383J. doi:10.2475/ajs.s5-35.209.383.
- Tedford, Richard H.; Wang, Xiaoming; Taylor, Beryl E. (2009). "Phylogenetic Systematics of the North American Fossil Caninae (Carnivora: Canidae)" (PDF). Bulletin of the American Museum of Natural History. 325: 1–218. doi:10.1206/574.1. hdl:2246/5999. S2CID 83594819.
- Tedford, R.H. & Qiu, Z.-X., 1996 - A new canid genus from the Pliocene of Yushe, Shanxi Province – Vertebrata PalAsiatica 34 (1): 27–40
- Anderson, E. (1996). "A preliminary report on the Carnivora of Porcupine Cave, Park County, Colorado". In Churcher, C. S. R.; Stewart, K. M.; Seymour, K. L. (eds.). Palaeoecology and palaeoenvironments of late Cenozoic mammals. Toronto: University of Toronto Press. pp. 259–282. ISBN 978-0-8020-0728-5.
- Albright, III, L. B. (2000). Biostratigraphy and Vertebrate Paleontology of the San Timoteo Badlands, Southern California. University of California Publications in Geological Sciences. 144. University of California Press. pp. 1–121. ISBN 978-0-520-91598-5.
- "Canis edwardii". Fossilworks. Retrieved 11 July 2016.
- Berta, A. (1995). "Fossil carnivores from the Leisey Shell Pits, Hillsborough County, Florida". In Hulbert, Jr., R. C.; Morgan, G. S.; Webb, S. D. (eds.). Paleontology and geology of the Leisey Shell Pits, early Pleistocene of Florida. 37. Bulletin of the Florida Museum of Natural History. pp. 463–499.
- "Canis ambrusteri". Fossilworks. Retrieved 11 July 2016.
- "Canis dirus". Fossilworks. Retrieved 11 July 2016.
- Kurten, B. 1984: Geographic differentiation in the Rancholabrean dire wolf (Canis dirus Leidy) in North America. In Genoways, H. H. & Dawson, M. R. (eds.): Contributions in Quaternary Vertebrate Paleontology: A Volume in Memorial to John E. Guilday, 218–227. Carnegie Museum of Natural History Special Publication 8
- Geist, Valerius (1998). Deer of the World: Their Evolution, Behaviour, and Ecology (1 ed.). Stackpole Books, Mechanicsburg, Pennsylvania. p. 10. ISBN 0811704963.
- Geist, Valerius (1987). "On speciation in Ice Age mammals, with special reference to cervids and caprids". Canadian Journal of Zoology. 65 (5): 1067–1084. doi:10.1139/z87-171.
- Goulet, G. D. (1993). Comparison of temporal and geographical skull variation among Nearctic, modern, Holocene, and late Pleistocene gray wolves (Canis lupus) and selected Canis (Master's Degree). Winnipeg: University of Manitoba. pp. 1–116.
- Berta, A. (1988). Quaternary evolution and biogeography of the large South American Canidae (Mammalia: Carnivora). 132. University of California Publications in Geological Sciences. pp. 1–49. ISBN 978-0-520-09960-9.
- Nowak, R. M.; Federoff, N. E. (2002). "The systematic status of the Italian wolf Canis lupus". Acta Theriol. 47 (3): 333–338. doi:10.1007/BF03194151. S2CID 366077.
- Canis occidentalis furlongi Merriam (1910), Univ. California Publ. Bull. Dept. Geol., 5:393
- Stevenson, Marc (1978). "9–Dire wolf Systematics and Behavior". In Hall, Roberta L.; Sharp, Henry S. (eds.). Wolf and Man: Evolution in Parallel. Academic Press. pp. 183–4. ISBN 0123192501.
- Canis milleri Merriam (1912) Mem. Univ. California, 1:247
- Merriam, J. C. (1912). "The fauna of Rancho La Brea, Part II. Canidae". Memoirs of the University of California. 1: 217–273.
- Aenocyon milleri Merriam (1918) Univ. California Publ. Bull. Dept. Geol., 10:533.
- Kurtén, B. (1968). Pleistocene mammals of Europe. London: Weidenfeld and Nicolson. p. 317. ISBN 978-0-202-30953-8.
- Tong; Hu, N.; Wang, X. (2012). "New remains of Canis chihliensis (Mammalia, Carnivora) from Shanshenmiaozui, a lower Pleistocene site in Yangyuan, Hebei". Vertebrata PalAsiatica. 50 (4): 335–360.
- Koler-Matznick, Janice (2002). "The origin of the dog revisited" (PDF). Anthrozoos: A Multidisciplinary Journal of the Interactions of People & Animals. 15 (2): 98–118. doi:10.2752/089279302786992595. S2CID 14850835.
- Olsen, S. J. (1985). Origins of the domestic dog: the fossil record. Univ. of Arizona Press, Tucson, USA. pp. 88–89.
- Olsen S J, Olsen J W, Qi G Q, 1982. The position of Canis lupus variabilis, from Zhoukoudian, in the ancestral lineage of the domestic dog, Canis familiaris. Vert PalAsia, 20(3): 264–267(in Chinese with last page abstract in English)
- Lawrence, B. 1967. Early domestic dogs. Zeitschrift für Säugetierkunde 32: 44 – 59.
- Pei, W.C. (1934). The carnivora from locality 1 of Choukoutien. Palaeontologia Sinica, Series C, vol. 8, Fascicle 1. Geological Survey of China, Beijing. pp. 1–45.
- Westgate, John A; Pearce, G. William; Preece, Shari J; Schweger, Charles E; Morlan, Richard E; Pearce, Nicholas J.G; Perkins, T. William (2017). "Tephrochronology, magnetostratigraphy and mammalian faunas of Middle and Early Pleistocene sediments at two sites on the Old Crow River, northern Yukon Territory, Canada". Quaternary Research. 79: 75–85. doi:10.1016/j.yqres.2012.09.003.
- Bonifay, M.F., 1971. Carnivores quaternaires du Sud-Est de la France. Muséum national d'Histoire naturelle, Paris, 334 p. (Mémoires du Muséum national d'Histoire naturelle, Sér. C – Sciences de la Terre (1950–1992) ; 21 (2)). [Quaternary carnivores of the South-East of France]
- Argant A., 1991 – Carnivores quaternaires de Bourgogne. Documents du Laboratoire de Géologie de Lyon, n° 115, 301 p.
- Boudadi-Maligne, Myriam (2012). "Une nouvelle sous-espèce de loup (Canis lupus maximus nov. Subsp.) dans le Pléistocène supérieur d'Europe occidentale [A new subspecies of wolf (Canis lupus maximus nov. subsp.) from the upper Pleistocene of Western Europe]". Comptes Rendus Palevol. 11 (7): 475. doi:10.1016/j.crpv.2012.04.003.
- Brugal, J. P.; Boudadi-Maligne, M. (2011). "Quaternary small to large canids in Europe: taxonomic status and biochronological contribution". Quaternary International. 243: 171–182. doi:10.1016/j.quaint.2011.01.046.
- Anzidei, Anna Paola; Bulgarelli, Grazia Maria; Catalano, Paola; Cerilli, Eugenio; Gallotti, Rosalia; Lemorini, Cristina; Milli, Salvatore; Palombo, Maria Rita; Pantano, Walter; Santucci, Ernesto (2012). "Ongoing research at the late Middle Pleistocene site of La Polledrara di Cecanibbio (central Italy), with emphasis on human–elephant relationships". Quaternary International. 255: 171–187. doi:10.1016/j.quaint.2011.06.005.
- Lucenti, Saverio Bartolini; Bukhsianidze, Maia; Martínez-Navarro, Bienvenido; Lordkipanidze, David (2020). "The Wolf from Dmanisi and Augmented Reality: Review, Implications, and Opportunities". Frontiers in Earth Science. 8: 131. Bibcode:2020FrEaS...8..131B. doi:10.3389/feart.2020.00131. S2CID 216337804.
- Thalmann, Olaf; Perri, Angela R. (2018). "Paleogenomic Inferences of Dog Domestication". In Lindqvist, C.; Rajora, O. (eds.). Paleogenomics. Population Genomics. Springer, Cham. pp. 273–306. doi:10.1007/13836_2018_27. ISBN 978-3-030-04752-8.
- Germonpré, Mietje; Sablin, Mikhail V.; Stevens, Rhiannon E.; Hedges, Robert E.M.; Hofreiter, Michael; Stiller, Mathias; Després, Viviane R. (2009). "Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes". Journal of Archaeological Science. 36 (2): 473–490. doi:10.1016/j.jas.2008.09.033.
- Sablin, Mikhail V.; Khlopachev, Gennady A. (2002). "The Earliest Ice Age Dogs:Evidence from Eliseevichi I" (PDF). Wenner-Gren Foundation for Anthropological Research. Retrieved 10 January 2015. Cite journal requires
|journal=(help) - Thalmann, O.; Shapiro, B.; Cui, P.; Schuenemann, V. J.; Sawyer, S. K.; Greenfield, D. L.; Germonpre, M. B.; Sablin, M. V.; Lopez-Giraldez, F.; Domingo-Roura, X.; Napierala, H.; Uerpmann, H.-P.; Loponte, D. M.; Acosta, A. A.; Giemsch, L.; Schmitz, R. W.; Worthington, B.; Buikstra, J. E.; Druzhkova, A.; Graphodatsky, A. S.; Ovodov, N. D.; Wahlberg, N.; Freedman, A. H.; Schweizer, R. M.; Koepfli, K.- P.; Leonard, J. A.; Meyer, M.; Krause, J.; Paabo, S.; et al. (2013). "Complete Mitochondrial Genomes of Ancient Canids Suggest a European Origin of Domestic Dogs". Science. 342 (6160): 871–4. Bibcode:2013Sci...342..871T. doi:10.1126/science.1243650. hdl:10261/88173. PMID 24233726. S2CID 1526260.
- Morey, Darcy F.; Jeger, Rujana (2015). "Paleolithic dogs: Why sustained domestication then?". Journal of Archaeological Science: Reports. 3: 420–428. doi:10.1016/j.jasrep.2015.06.031.
- Wayne, Robert K. (1986). "Cranial Morphology of Domestic and Wild Canids: The Influence of Development on Morphological Change". Evolution. 40 (2): 243–261. doi:10.2307/2408805. JSTOR 2408805. PMID 28556057.
- Drake, Abby Grace; Klingenberg, Christian Peter (2010). "Large‐Scale Diversification of Skull Shape in Domestic Dogs: Disparity and Modularity". The American Naturalist. 175 (3): 289–301. doi:10.1086/650372. PMID 20095825. S2CID 26967649.
- Roberts, Taryn; McGreevy, Paul; Valenzuela, Michael (2010). "Human Induced Rotation and Reorganization of the Brain of Domestic Dogs". PLOS ONE. 5 (7): e11946. doi:10.1371/journal.pone.0011946. PMC 2909913. PMID 20668685.
- Drake, Abby Grace, "Evolution and development of the skull morphology of canids: An investigation of morphological integration and heterochrony" (January 1, 2004). Doctoral Dissertations Available from Proquest. Paper AAI3136721. link
- Drake, Abby Grace (2011). "Dispelling dog dogma: An investigation of heterochrony in dogs using 3D geometric morphometric analysis of skull shape". Evolution & Development. 13 (2): 204–213. doi:10.1111/j.1525-142X.2011.00470.x. PMID 21410876. S2CID 20893501.
- Perri, Angela (2016). "A wolf in dog's clothing: Initial dog domestication and Pleistocene wolf variation". Journal of Archaeological Science. 68: 1–4. doi:10.1016/j.jas.2016.02.003.
- Rand, David M. (2001). "The Units of Selection on Mitochondrial DNA". Annual Review of Ecology and Systematics. 32: 415–448. doi:10.1146/annurev.ecolsys.32.081501.114109.
- Robert K. Wayne, Jennifer A. Leonard, Carles Vila (2006). "Chapter 19:Genetic Analysis of Dog Domestication". In Melinda A. Zeder (ed.). Documenting Domestication:New Genetic and Archaeological Paradigms. University of California Press. pp. 279–295. ISBN 978-0-520-24638-6.CS1 maint: uses authors parameter (link)
- Hofreiter, Michael; Serre, David; Poinar, Hendrik N.; Kuch, Melanie; Pääbo, Svante (2001). "Ancient DNA". Nature Reviews Genetics. 2 (5): 353–9. doi:10.1038/35072071. PMID 11331901. S2CID 205016024.
- Li, Wen-Hsiung (1997). Molecular evolution. Sunderland, Massachusetts: Sinauer Associates. ISBN 978-0-87893-463-8.
- Pesole, G; Gissi, C; De Chirico, A; Saccone, C (1999). "Nucleotide substitution rate of mammalian mitochondrial genomes". Journal of Molecular Evolution. 48 (4): 427–34. doi:10.1007/pl00006487. PMID 10079281. S2CID 1665621.
- Wan, Qiu-Hong; Wu, Hua; Fujihara, Tsutomu; Fang, Sheng-Guo (2003). "Which genetic marker for which conservation genetics issue?" (PDF). Electrophoresis. 25 (14): 2165–2176. doi:10.1002/elps.200305922. PMID 15274000. S2CID 11567406.
- Birky, C. William (2001). "The Inheritance of Genes in Mitochondria and Chloroplasts: Laws, Mechanisms, and Models". Annual Review of Genetics. 35: 125–48. doi:10.1146/annurev.genet.35.102401.090231. PMID 11700280.
- Avise, J. C. (1994). Molecular Markers, Natural History, and Evolution. New York: Chapman & Hall. ISBN 978-0-412-03781-8.
- Avise, J. C. (2000). Phylogeography: The History and Formation of Species. Cambridge, Massachusetts: Harvard University Press. ISBN 978-0-674-66638-2.
- Lindblad-Toh, Kerstin; Wade, Claire M.; Mikkelsen, Tarjei S.; Karlsson, Elinor K.; Jaffe, David B.; Kamal, Michael; Clamp, Michele; Chang, Jean L.; Kulbokas, Edward J.; Zody, Michael C.; Mauceli, Evan; Xie, Xiaohui; Breen, Matthew; Wayne, Robert K.; Ostrander, Elaine A.; Ponting, Chris P.; Galibert, Francis; Smith, Douglas R.; Dejong, Pieter J.; Kirkness, Ewen; Alvarez, Pablo; Biagi, Tara; Brockman, William; Butler, Jonathan; Chin, Chee-Wye; Cook, April; Cuff, James; Daly, Mark J.; Decaprio, David; et al. (2005). "Genome sequence, comparative analysis and haplotype structure of the domestic dog". Nature. 438 (7069): 803–819. Bibcode:2005Natur.438..803L. doi:10.1038/nature04338. PMID 16341006.
- Koepfli, K.-P.; Pollinger, J.; Godinho, R.; Robinson, J.; Lea, A.; Hendricks, S.; Schweizer, R. M.; Thalmann, O.; Silva, P.; Fan, Z.; Yurchenko, A. A.; Dobrynin, P.; Makunin, A.; Cahill, J. A.; Shapiro, B.; Álvares, F.; Brito, J. C.; Geffen, E.; Leonard, J. A.; Helgen, K. M.; Johnson, W. E.; O’Brien, S. J.; Van Valkenburgh, B.; Wayne, R. K. (2015-08-17). "Genome-wide Evidence Reveals that African and Eurasian Golden Jackals Are Distinct Species". Current Biology. 25 (16): 2158–65. doi:10.1016/j.cub.2015.06.060. PMID 26234211.
- Vila, C. (1997). "Multiple and ancient origins of the domestic dog". Science. 276 (5319): 1687–9. doi:10.1126/science.276.5319.1687. PMID 9180076.
- Wayne, R. & Ostrander, Elaine A. (1999). "Origin, genetic diversity, and genome structure of the domestic dog". BioEssays. 21 (3): 247–57. doi:10.1002/(SICI)1521-1878(199903)21:3<247::AID-BIES9>3.0.CO;2-Z. PMID 10333734.CS1 maint: uses authors parameter (link)
- Boyko, A. (2009). "Complex population structure in African village dogs and its implications for inferring dog domestication history". Proceedings of the National Academy of Sciences. 106 (33): 13903–13908. Bibcode:2009PNAS..10613903B. doi:10.1073/pnas.0902129106. PMC 2728993. PMID 19666600.
- Pang, J. (2009). "mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves". Molecular Biology and Evolution. 26 (12): 2849–64. doi:10.1093/molbev/msp195. PMC 2775109. PMID 19723671.
- Cronin, M. A.; Canovas, A.; Bannasch, D. L.; Oberbauer, A. M.; Medrano, J. F. (2014). "Single Nucleotide Polymorphism (SNP) Variation of Wolves (Canis lupus) in Southeast Alaska and Comparison with Wolves, Dogs, and Coyotes in North America". Journal of Heredity. 106 (1): 26–36. doi:10.1093/jhered/esu075. PMID 25429025.
- Bardeleben, Carolyne; Moore, Rachael L.; Wayne, Robert K. (2005). "A molecular phylogeny of the Canidae based on six nuclear loci". Molecular Phylogenetics and Evolution. 37 (3): 815–31. doi:10.1016/j.ympev.2005.07.019. PMID 16213754.
- Gray, Melissa M; Sutter, Nathan B; Ostrander, Elaine A; Wayne, Robert K (2010). "The IGF1 small dog haplotype is derived from Middle Eastern grey wolves". BMC Biology. 8: 16. doi:10.1186/1741-7007-8-16. PMC 2837629. PMID 20181231.
- Wayne, Robert K.; Vonholdt, Bridgett M. (2012). "Evolutionary genomics of dog domestication". Mammalian Genome. 23 (1–2): 3–18. doi:10.1007/s00335-011-9386-7. PMID 22270221. S2CID 16003335.
- Mech, L. David; Barber-meyer, Shannon (2017). "Use of erroneous wolf generation time in assessments of domestic dog and human evolution" (PDF). Science. 352: 1228–31. Bibcode:2016Sci...352.1228F. doi:10.1126/science.aaf3161. PMID 27257259. S2CID 206647686.
- Frantz, L. A. F.; Mullin, V. E.; Pionnier-Capitan, M.; Lebrasseur, O.; Ollivier, M.; Perri, A.; Linderholm, A.; Mattiangeli, V.; Teasdale, M. D.; Dimopoulos, E. A.; Tresset, A.; Duffraisse, M.; McCormick, F.; Bartosiewicz, L.; Gal, E.; Nyerges, E. A.; Sablin, M. V.; Brehard, S.; Mashkour, M.; b l Escu, A.; Gillet, B.; Hughes, S.; Chassaing, O.; Hitte, C.; Vigne, J.-D.; Dobney, K.; Hanni, C.; Bradley, D. G.; Larson, G. (2016). "Genomic and archaeological evidence suggest a dual origin of domestic dogs". Science. 352 (6290): 1228–31. Bibcode:2016Sci...352.1228F. doi:10.1126/science.aaf3161. PMID 27257259. S2CID 206647686.
- Wayne, R. (1993). "Molecular evolution of the dog family". Trends in Genetics. 9 (6): 218–24. doi:10.1016/0168-9525(93)90122-X. PMID 8337763.
- Wayne, R. K.; Benvenista, R. E.; Janczewski, D. N.; O'Brien, S. J. (1989). "17-Molecular and biomechanical evolution of the Carnivora". In Gittleman, J. L. (ed.). Carnivore Behavior, Ecology, and Evolution. Springer. pp. 477–478. ISBN 9780412343605.
- Vila, C.; Leonard, J. (2012). "1-Canid Phylogeny and the Origin of the Domestic Dog". In Ostrander, E. A.; Ruvinski, A. (eds.). Genetics of the Dog (2 ed.). CABI. p. 3. ISBN 9781845939403.
- Wurster-Hill, D. H.; Centerwall, W. R. (1982). "The interrelationships of chromosome banding patterns in canids, mustelids, hyena, and felids". Cytogenetics and Cell Genetics. 34 (1–2): 178–192. doi:10.1159/000131806. PMID 7151489.
- Gottelli, D.; Sillero-Zubiri, C.; Applebaum, G. D.; Roy, M. S.; Girman, D. J.; Garcia-Moreno, J.; Ostrander, E. A.; Wayne, R. K. (1994). "Molecular genetics of the most endangered canid: The Ethiopian wolf Canis simensis". Molecular Ecology. 3 (4): 301–12. doi:10.1111/j.1365-294X.1994.tb00070.x. PMID 7921357. S2CID 25976335.
- Pocock, R. I. (1941). Fauna of British India: Mammals. 2. Taylor & Francis. pp. 146–163.
- Girman, D. J.; Vilà, C.; Geffen, E.; Creel, S.; Mills, M. G. L.; McNutt, J. W.; Ginsberg, J.; Kat, P. W.; Mamiya, K. H.; Wayne, R. K. (2001). "Patterns of population subdivision, gene flow and genetic variability in the African wild dog (Lycaon pictus)". Molecular Ecology. 10 (7): 1703–23. doi:10.1046/j.0962-1083.2001.01302.x. PMID 11472538. S2CID 24540274.
- Wayne, R.K.; Meyer, A.; Lehman, N.; van Valkenburgh, B.; Kat, P.W.; Fuller, T.K.; Girman, D.; O'Brien, S.J. (1990). "Large sequence divergence among mitochondrial DNA genotypes within populations of eastern African black-backed jackals" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 87 (5): 1772–1776. Bibcode:1990PNAS...87.1772W. doi:10.1073/pnas.87.5.1772. PMC 53565. PMID 1968637. Retrieved 21 December 2011.
- Juliane Kaminski and Sarah Marshall-Pescini (2014). "Chapter 1 - The Social Dog:History and Evolution". The Social Dog:Behavior and Cognition. Elsevier. p. 4. ISBN 978-0-12407-931-1.CS1 maint: uses authors parameter (link)
- Gopalakrishnan, Shyam; Sinding, Mikkel-Holger S.; Ramos-Madrigal, Jazmín; Niemann, Jonas; Samaniego Castruita, Jose A.; Vieira, Filipe G.; Carøe, Christian; Montero, Marc de Manuel; Kuderna, Lukas; Serres, Aitor; González-Basallote, Víctor Manuel; Liu, Yan-Hu; Wang, Guo-Dong; Marques-Bonet, Tomas; Mirarab, Siavash; Fernandes, Carlos; Gaubert, Philippe; Koepfli, Klaus-Peter; Budd, Jane; Rueness, Eli Knispel; Heide-Jørgensen, Mads Peter; Petersen, Bent; Sicheritz-Ponten, Thomas; Bachmann, Lutz; Wiig, Øystein; Hansen, Anders J.; Gilbert, M. Thomas P. (2018). "Interspecific Gene Flow Shaped the Evolution of the Genus Canis". Current Biology. 28 (21): 3441–3449.e5. doi:10.1016/j.cub.2018.08.041. PMC 6224481. PMID 30344120.
- Ersmark, Erik; Klütsch, Cornelya F. C.; Chan, Yvonne L.; Sinding, Mikkel-Holger S.; Fain, Steven R.; Illarionova, Natalia A.; Oskarsson, Mattias; Uhlén, Mathias; Zhang, Ya-Ping; Dalén, Love; Savolainen, Peter (2016). "From the Past to the Present: Wolf Phylogeography and Demographic History Based on the Mitochondrial Control Region". Frontiers in Ecology and Evolution. 4. doi:10.3389/fevo.2016.00134.
- Werhahn, Geraldine; Senn, Helen; Kaden, Jennifer; Joshi, Jyoti; Bhattarai, Susmita; Kusi, Naresh; Sillero-Zubiri, Claudio; MacDonald, David W. (2017). "Phylogenetic evidence for the ancient Himalayan wolf: Towards a clarification of its taxonomic status based on genetic sampling from western Nepal". Royal Society Open Science. 4 (6): 170186. Bibcode:2017RSOS....470186W. doi:10.1098/rsos.170186. PMC 5493914. PMID 28680672.
- Werhahn, Geraldine; Senn, Helen; Ghazali, Muhammad; Karmacharya, Dibesh; Sherchan, Adarsh Man; Joshi, Jyoti; Kusi, Naresh; López-Bao, José Vincente; Rosen, Tanya; Kachel, Shannon; Sillero-Zubiri, Claudio; MacDonald, David W. (2018). "The unique genetic adaptation of the Himalayan wolf to high-altitudes and consequences for conservation". Global Ecology and Conservation. 16: e00455. doi:10.1016/j.gecco.2018.e00455.
- Aggarwal, R. K.; Kivisild, T.; Ramadevi, J.; Singh, L. (2007). "Mitochondrial DNA coding region sequences support the phylogenetic distinction of two Indian wolf species". Journal of Zoological Systematics and Evolutionary Research. 45 (2): 163–172. doi:10.1111/j.1439-0469.2006.00400.x.
- Sharma, D. K.; Maldonado, J. E.; Jhala, Y. V.; Fleischer, R. C. (2004). "Ancient wolf lineages in India". Proceedings of the Royal Society B: Biological Sciences. 271 (Suppl 3): S1–S4. doi:10.1098/rsbl.2003.0071. PMC 1809981. PMID 15101402.
- Leonard, J. A.; Vilà, C; Fox-Dobbs, K; Koch, P. L.; Wayne, R. K.; Van Valkenburgh, B (2007). "Megafaunal extinctions and the disappearance of a specialized wolf ecomorph" (PDF). Current Biology. 17 (13): 1146–50. doi:10.1016/j.cub.2007.05.072. PMID 17583509. S2CID 14039133.
- Pilot, M.; et al. (2010). "Phylogeographic history of grey wolves in Europe". BMC Evolutionary Biology. 10: 104. doi:10.1186/1471-2148-10-104. PMC 2873414. PMID 20409299.
- Rueness, Eli Knispel; Asmyhr, Maria Gulbrandsen; Sillero-Zubiri, Claudio; MacDonald, David W; Bekele, Afework; Atickem, Anagaw; Stenseth, Nils Chr (2011). "The Cryptic African Wolf: Canis aureus lupaster is Not a Golden Jackal and is Not Endemic to Egypt". PLOS ONE. 6 (1): e16385. Bibcode:2011PLoSO...616385R. doi:10.1371/journal.pone.0016385. PMC 3027653. PMID 21298107.
- Miklosi, Adam (2015). Dog Behaviour, Evolution, and Cognition. Oxford Biology (2 ed.). Oxford University Press. pp. 106–107. ISBN 978-0199545667.
- Loog, Liisa; Thalmann, Olaf; Sinding, Mikkel‐Holger S.; Schuenemann, Verena J.; Perri, Angela; Germonpré, Mietje; Bocherens, Herve; Witt, Kelsey E.; Samaniego Castruita, Jose A.; Velasco, Marcela S.; Lundstrøm, Inge K.C.; Wales, Nathan; Sonet, Gontran; Frantz, Laurent; Schroeder, Hannes; Budd, Jane; Jimenez, Elodie‐Laure; Fedorov, Sergey; Gasparyan, Boris; Kandel, Andrew W.; Lázničková‐Galetová, Martina; Napierala, Hannes; Uerpmann, Hans‐Peter; Nikolskiy, Pavel A.; Pavlova, Elena Y.; Pitulko, Vladimir V.; Herzig, Karl‐Heinz; Malhi, Ripan S.; Willerslev, Eske; et al. (2019). "Ancient DNA suggests modern wolves trace their origin to a late Pleistocene expansion from Beringia". Molecular Ecology. 29 (9): 1596–1610. doi:10.1111/mec.15329. PMC 7317801. PMID 31840921.
- Koblmüller, Stephan; Vilà, Carles; Lorente-Galdos, Belen; Dabad, Marc; Ramirez, Oscar; Marques-Bonet, Tomas; Wayne, Robert K.; Leonard, Jennifer A. (2016). "Whole mitochondrial genomes illuminate ancient intercontinental dispersals of grey wolves (Canis lupus)". Journal of Biogeography. 43 (9): 1728–1738. doi:10.1111/jbi.12765. hdl:10261/153364.
- Werhahn, Geraldine; Senn, Helen; Ghazali, Muhammad; Karmacharya, Dibesh; Sherchan, Adarsh Man; Joshi, Jyoti; Kusi, Naresh; López-Bao, José Vincente; Rosen, Tanya; Kachel, Shannon; Sillero-Zubiri, Claudio; MacDonald, David W. (2018). "The unique genetic adaptation of the Himalayan wolf to high-altitudes and consequences for conservation". Global Ecology and Conservation. 16: e00455. doi:10.1016/j.gecco.2018.e00455.
- Schweizer, Rena M.; Wayne, Robert K. (2020). "Illuminating the mysteries of wolf history". Molecular Ecology. 29 (9): 1589–1591. doi:10.1111/MEC.15438. PMID 32286714.
- Wang, Guo-Dong; Zhang, Ming; Wang, Xuan; Yang, Melinda A.; Cao, Peng; Liu, Feng; Lu, Heng; Feng, Xiaotian; Skoglund, Pontus; Wang, Lu; Fu, Qiaomei; Zhang, Ya-Ping (2019). "Genomic Approaches Reveal an Endemic Subpopulation of Gray Wolves in Southern China". iScience. 20: 110–118. doi:10.1016/j.isci.2019.09.008. PMC 6817678. PMID 31563851.
- Cox, C. B.; Moore, Peter D.; Ladle, Richard (2016). Biogeography: An Ecological and Evolutionary Approach. Wiley-Blackwell. p. 106. ISBN 978-1-118-96858-1.
- Editorial Board (2012). Concise Dictionary of Science. New Delhi: V&S Publishers. p. 137. ISBN 978-93-81588-64-2.
- Arora, Devender; Singh, Ajeet; Sharma, Vikrant; Bhaduria, Harvendra Singh; Patel, Ram Bahadur (2015). "Hgs Db: Haplogroups Database to understand migration and molecular risk assessment". Bioinformation. 11 (6): 272–5. doi:10.6026/97320630011272. PMC 4512000. PMID 26229286.
- "Genetics Glossary". International Society of Genetic Genealogy. 2015. Retrieved 22 July 2016.
- Randi, Ettore (2011). "Genetics and conservation of wolves Canis lupus in Europe". Mammal Review. 41 (2): 99–111. doi:10.1111/j.1365-2907.2010.00176.x.
- Tamm, E.; Kivisild, T.; Reidla, M.; Metspalu, M.; Smith, D. G.; Mulligan, C. J.; Bravi, C. M.; Rickards, O.; Martinez-Labarga, C.; Khusnutdinova, E. K.; Fedorova, S. A.; Golubenko, M. V.; Stepanov, V. A.; Gubina, M. A.; Zhadanov, S. I.; Ossipova, L. P.; Damba, L.; Voevoda, M. I.; Dipierri, J. E.; Villems, R.; Malhi, R. S. (2007). Carter, Dee (ed.). "Beringian Standstill and Spread of Native American Founders". PLOS ONE. 2 (9): e829. Bibcode:2007PLoSO...2..829T. doi:10.1371/journal.pone.0000829. PMC 1952074. PMID 17786201.
- Hofreiter, Michael; Barnes, Ian (2010). "Diversity lost: Are all Holarctic large mammal species just relict populations?". BMC Biology. 8: 46. doi:10.1186/1741-7007-8-46. PMC 2858106. PMID 20409351.
- Germonpré, Mietje; Sablin, Mikhail V.; Després, Viviane; Hofreiter, Michael; Lázničková-Galetová, Martina; Stevens, Rhiannon E.; Stiller, Mathias (2013). "Palaeolithic dogs and the early domestication of the wolf: A reply to the comments of Crockford and Kuzmin (2012)". Journal of Archaeological Science. 40: 786–792. doi:10.1016/j.jas.2012.06.016.
- Hofreiter, Michael (2007). "Pleistocene Extinctions: Haunting the Survivors". Current Biology. 17 (15): R609–11. doi:10.1016/j.cub.2007.06.031. PMID 17686436.
- Pilot, M; Greco, C; Vonholdt, B M; Jędrzejewska, B; Randi, E; Jędrzejewski, W; Sidorovich, V E; Ostrander, E A; Wayne, R K (2013). "Genome-wide signatures of population bottlenecks and diversifying selection in European wolves". Heredity. 112 (4): 428–42. doi:10.1038/hdy.2013.122. PMC 3966127. PMID 24346500.
- Weckworth, Byron V.; Talbot, Sandra; Sage, George K.; Person, David K.; Cook, Joseph (2005). "A Signal for Independent Coastal and Continental histories among North American wolves". Molecular Ecology. 14 (4): 917–31. doi:10.1111/j.1365-294X.2005.02461.x. PMID 15773925. S2CID 12896064.
- Weckworth, Byron V.; Talbot, Sandra L.; Cook, Joseph A. (2010). "Phylogeography of wolves (Canis lupus) in the Pacific Northwest". Journal of Mammalogy. 91 (2): 363–375. doi:10.1644/09-MAMM-A-036.1.
- Weckworth, Byron V.; Dawson, Natalie G.; Talbot, Sandra L.; Flamme, Melanie J.; Cook, Joseph A. (2011). "Going Coastal: Shared Evolutionary History between Coastal British Columbia and Southeast Alaska Wolves (Canis lupus)". PLOS ONE. 6 (5): e19582. Bibcode:2011PLoSO...619582W. doi:10.1371/journal.pone.0019582. PMC 3087762. PMID 21573241.
- Vonholdt, B. M.; Cahill, J. A.; Fan, Z.; Gronau, I.; Robinson, J.; Pollinger, J. P.; Shapiro, B.; Wall, J.; Wayne, R. K. (2016). "Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf". Science Advances. 2 (7): e1501714. Bibcode:2016SciA....2E1714V. doi:10.1126/sciadv.1501714. PMC 5919777. PMID 29713682.
- Matsumura, Shuichi; Inoshima, Yasuo; Ishiguro, Naotaka (2014). "Reconstructing the colonization history of lost wolf lineages by the analysis of the mitochondrial genome". Molecular Phylogenetics and Evolution. 80: 105–12. doi:10.1016/j.ympev.2014.08.004. PMID 25132126.
- Abe, H. (1999). "Diversity and conservation of mammals of Japan". In Yokohata, Y.; Nakamura, S. (eds.). Recent Advances in the Biology of Japanese Insectivore. Shōbara: Hiba Society of Natural History. pp. 89–104.
- Ishiguro, Naotaka; Inoshima, Yasuo; Shigehara, Nobuo; Ichikawa, Hideo; Kato, Masaru (2010). "Osteological and Genetic Analysis of the Extinct Ezo Wolf (Canis Lupus Hattai) from Hokkaido Island, Japan". Zoological Science. 27 (4): 320–4. doi:10.2108/zsj.27.320. PMID 20377350. S2CID 11569628.
- Walker, Brett (2008). The Lost Wolves of Japan. University of Washington Press. p. 42.
- Ishiguro, Naotaka; Inoshima, Yasuo; Shigehara, Nobuo (2009). "Mitochondrial DNA Analysis of the Japanese Wolf (Canis lupus hodophilax Temminck, 1839) and Comparison with Representative Wolf and Domestic Dog Haplotypes". Zoological Science. 26 (11): 765–70. doi:10.2108/zsj.26.765. PMID 19877836. S2CID 27005517.
- Pardi, Melissa I.; Smith, Felisa A. (2016). "Biotic responses of canids to the terminal Pleistocene megafauna extinction". Ecography. 39 (2): 141–151. doi:10.1111/ecog.01596.
- Witt, Kelsey E.; Judd, Kathleen; Kitchen, Andrew; Grier, Colin; Kohler, Timothy A.; Ortman, Scott G.; Kemp, Brian M.; Malhi, Ripan S. (2015). "DNA analysis of ancient dogs of the Americas: Identifying possible founding haplotypes and reconstructing population histories". Journal of Human Evolution. 79: 105–18. doi:10.1016/j.jhevol.2014.10.012. PMID 25532803.
- Wolf and Man. Evolution in Parallel. R.L. Hall and H.S. Sharp. Academic Press, New York, 1978
- Leonard, Jennifer A.; Vilà, Carles; Wayne, Robert K. (2004). "Fast Track: Legacy lost: Genetic variability and population size of extirpated US grey wolves (Canis lupus)". Molecular Ecology. 14 (1): 9–17. doi:10.1111/j.1365-294X.2004.02389.x. PMID 15643947. S2CID 11343074.
- Meachen, Julie; Wooller, Matthew J.; Barst, Benjamin D.; Funck, Juliette; Crann, Carley; Heath, Jess; Cassatt-Johnstone, Molly; Shapiro, Beth; Hall, Elizabeth; Hewitson, Susan; Zazula, Grant (2020). "A mummified Pleistocene gray wolf pup". Current Biology. 30 (24): R1467–R1468. doi:10.1016/j.cub.2020.11.011.
- Morell, Virginia (2016). "How do you save a wolf that's not really a wolf?". Science. 353 (6300). doi:10.1126/science.aag0699.
- Vila, C.; Amorim, I. R.; Leonard, J. A.; Posada, D.; Castroviejo, J.; Petrucci-Fonseca, F.; Crandall, K. A.; Ellegren, H.; Wayne, R. K. (1999). "Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus" (PDF). Molecular Ecology. 8 (12): 2089–103. doi:10.1046/j.1365-294x.1999.00825.x. hdl:10261/58565. PMID 10632860. S2CID 31680635.
- Larson G, Bradley DG (2014). "How Much Is That in Dog Years? The Advent of Canine Population Genomics". PLOS Genetics. 10 (1): e1004093. doi:10.1371/journal.pgen.1004093. PMC 3894154. PMID 24453989.CS1 maint: uses authors parameter (link)
- Morey, Darcy F.; Jeger, Rujana (2016). "From wolf to dog: Late Pleistocene ecological dynamics, altered trophic strategies, and shifting human perceptions". Historical Biology. 29 (7): 1–9. doi:10.1080/08912963.2016.1262854. S2CID 90588969.
- Van Valkenburgh, Blaire; Hayward, Matthew W.; Ripple, William J.; Meloro, Carlo; Roth, V. Louise (2016). "The impact of large terrestrial carnivores on Pleistocene ecosystems". Proceedings of the National Academy of Sciences. 113 (4): 862–867. doi:10.1073/pnas.1502554112. PMC 4743832. PMID 26504224.
- Shipman, P. (2015). The Invaders:How humans and their dogs drove Neanderthals to extinction. Harvard University Press. ISBN 978-0-674-73676-4.
- Cagan, Alex; Blass, Torsten (2016). "Identification of genomic variants putatively targeted by selection during dog domestication". BMC Evolutionary Biology. 16: 10. doi:10.1186/s12862-015-0579-7. PMC 4710014. PMID 26754411.
- Song, Jiao-Jiao; Wang, Wen-Zhi; Otecko, Newton O.; Peng, Min-Sheng; Zhang, Ya-Ping (2016). "Reconciling the conflicts between mitochondrial DNA haplogroup trees of Canis lupus". Forensic Science International: Genetics. 23: 83–85. doi:10.1016/j.fsigen.2016.03.008. PMID 27042801.
- Pilot, Małgorzata; Greco, Claudia; Vonholdt, Bridgett M; Randi, Ettore; Jędrzejewski, Włodzimierz; Sidorovich, Vadim E; Konopiński, Maciej K; Ostrander, Elaine A; Wayne, Robert K (2018). "Widespread, long-term admixture between grey wolves and domestic dogs across Eurasia and its implications for the conservation status of hybrids". Evolutionary Applications. 11 (5): 662–680. doi:10.1111/eva.12595. PMC 5978975. PMID 29875809.
- Greig, Karen; Boocock, James; Prost, Stefan; Horsburgh, K. Ann; Jacomb, Chris; Walter, Richard; Matisoo-Smith, Elizabeth (2015). "Complete Mitochondrial Genomes of New Zealand's First Dogs". PLOS ONE. 10 (10): e0138536. Bibcode:2015PLoSO..1038536G. doi:10.1371/journal.pone.0138536. PMC 4596854. PMID 26444283.
- Savolainen, P.; Leitner, T.; Wilton, A. N.; Matisoo-Smith, E.; Lundeberg, J. (2004). "A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA". Proceedings of the National Academy of Sciences. 101 (33): 12387–12390. Bibcode:2004PNAS..10112387S. doi:10.1073/pnas.0401814101. PMC 514485. PMID 15299143.
- Oskarsson, M. C. R.; Klutsch, C. F. C.; Boonyaprakob, U.; Wilton, A.; Tanabe, Y.; Savolainen, P. (2011). "Mitochondrial DNA data indicate an introduction through Mainland Southeast Asia for Australian dingoes and Polynesian domestic dogs". Proceedings of the Royal Society B: Biological Sciences. 279 (1730): 967–974. doi:10.1098/rspb.2011.1395. PMC 3259930. PMID 21900326.
- Ardalan, Arman; Oskarsson, Mattias; Natanaelsson, Christian; Wilton, Alan N.; Ahmadian, Afshin; Savolainen, Peter (2012). "Narrow genetic basis for the Australian dingo confirmed through analysis of paternal ancestry". Genetica. 140 (1–3): 65–73. doi:10.1007/s10709-012-9658-5. PMC 3386486. PMID 22618967.
- Gross, Michael (2015). "Are dogs just like us?". Current Biology. 25 (17): R733–6. doi:10.1016/j.cub.2015.08.018. PMID 26561653.
- "Ancient wolf genome pushes back dawn of the dog". Nature. 2015.
- Machugh, David E.; Larson, Greger; Orlando, Ludovic (2016). "Taming the Past: Ancient DNA and the Study of Animal Domestication". Annual Review of Animal Biosciences. 5: 329–351. doi:10.1146/annurev-animal-022516-022747. PMID 27813680.
- Ersmark, E (2016). "Large carnivore population turnover and ecological change during the Late Quaternary". Stockholm University. Phd Thesis under J. Leonard and L. Dalen
- Lee, E. (2015). "Ancient DNA analysis of the oldest canid species from the Siberian Arctic and genetic contribution to the domestic dog". PLOS ONE. 10 (5): e0125759. Bibcode:2015PLoSO..1025759L. doi:10.1371/journal.pone.0125759. PMC 4446326. PMID 26018528.
- Irizarry, Kristopher J. L.; Vasconcelos, Elton J. R. (2018). "Population Genomics of Domestication and Breed Development in Canines in the Context of Cognitive, Social, Behavioral, and Disease Traits". In Rajora, O. (ed.). Population Genomics. pp. 755–806. doi:10.1007/13836_2018_43. ISBN 978-3-030-04587-6.
- Bocherens, Hervé; Drucker, Dorothée G.; Bonjean, Dominique; Bridault, Anne; Conard, Nicholas J.; Cupillard, Christophe; Germonpré, Mietje; Höneisen, Markus; Münzel, Susanne C.; Napierala, Hannes; Patou-Mathis, Marylène; Stephan, Elisabeth; Uerpmann, Hans-Peter; Ziegler, Reinhard (2011). "Isotopic evidence for dietary ecology of cave lion (Panthera spelaea) in North-Western Europe: Prey choice, competition and implications for extinction". Quaternary International. 245 (2): 249–261. Bibcode:2011QuInt.245..249B. doi:10.1016/j.quaint.2011.02.023.
- Yeakel, J. D.; Guimaraes, P. R.; Bocherens, H.; Koch, P. L. (2013). "The impact of climate change on the structure of Pleistocene food webs across the mammoth steppe". Proceedings of the Royal Society B: Biological Sciences. 280 (1762): 20130239. doi:10.1098/rspb.2013.0239. PMC 3673045. PMID 23658198.
- Bocherens, Hervé (2015). "Isotopic tracking of large carnivore palaeoecology in the mammoth steppe". Quaternary Science Reviews. 117: 42–71. doi:10.1016/j.quascirev.2015.03.018.
- Dinnis, R.; Pate, A.; Reynolds, N. (2016). "Mid-to-late Marine Isotope Stage 3 mammal faunas of Britain: A new look". Proceedings of the Geologists' Association. 127 (4): 435–444. doi:10.1016/j.pgeola.2016.05.002.
- Boitani,L. (2003). "Wolf conservation and recovery" in Wolves:Behavior Ecology and Conservation, eds L.D. Mech and L. Boitani (Chicago, IL:Chicago University Press), 317–340. doi:10.7208/chicago/9780226516981.001.0001
- Leonard, Jennifer A.; Vilà, Carles; Wayne, Robert K. (2004). "FAST TRACK: Legacy lost: Genetic variability and population size of extirpated US grey wolves (Canis lupus)". Molecular Ecology. 14 (1): 9–17. doi:10.1111/j.1365-294X.2004.02389.x. PMID 15643947. S2CID 11343074.
- Vonholdt, Bridgett M.; Pollinger, John P.; Earl, Dent A.; Knowles, James C.; Boyko, Adam R.; Parker, Heidi; Geffen, Eli; Pilot, Malgorzata; Jedrzejewski, Wlodzimierz; Jedrzejewska, Bogumila; Sidorovich, Vadim; Greco, Claudia; Randi, Ettore; Musiani, Marco; Kays, Roland; Bustamante, Carlos D.; Ostrander, Elaine A.; Novembre, John; Wayne, Robert K. (2011). "A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids". Genome Research. 21 (8): 1294–305. doi:10.1101/gr.116301.110. PMC 3149496. PMID 21566151.
- Vonholdt, Bridgett M.; Pollinger, John P.; Earl, Dent A.; Parker, Heidi G.; Ostrander, Elaine A.; Wayne, Robert K. (2012). "Identification of recent hybridization between gray wolves and domesticated dogs by SNP genotyping". Mammalian Genome. 24 (1–2): 80–8. doi:10.1007/s00335-012-9432-0. PMID 23064780. S2CID 1161310.
- Ernst Mayr (1999). "VIII-Nongeographic speciation". Systematics and the Origin of Species, from the Viewpoint of a Zoologist. Harvard University Press. pp. 194–195. ISBN 9780674862500.
- Vila, C.; Sundqvist, A.-K.; Flagstad, O.; Seddon, J.; Rnerfeldt, S. B.; Kojola, I.; Casulli, A.; Sand, H.; Wabakken, P.; Ellegren, H. (2003). "Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant". Proceedings of the Royal Society B: Biological Sciences. 270 (1510): 91–97. doi:10.1098/rspb.2002.2184. PMC 1691214. PMID 12590776.
- Muñoz-Fuentes, Violeta; Darimont, Chris T.; Wayne, Robert K.; Paquet, Paul C.; Leonard, Jennifer A. (2009). "Ecological factors drive differentiation in wolves from British Columbia". Journal of Biogeography. 36 (8): 1516–1531. doi:10.1111/j.1365-2699.2008.02067.x. hdl:10261/61670.
- Valière, Nathaniel; Fumagalli, Luca; Gielly, Ludovic; Miquel, Christian; Lequette, Benoît; Poulle, Marie-Lazarine; Weber, Jean-Marc; Arlettaz, Raphaël; Taberlet, Pierre (2003). "Long-distance wolf recolonization of France and Switzerland inferred from non-invasive genetic sampling over a period of 10 years". Animal Conservation. 6: 83–92. doi:10.1017/S1367943003003111.
- Fabbri, E.; Caniglia, R.; Kusak, J.; Galov, A.; Gomerčić, T.; Arbanasić, H.; Huber, D.; Randi, E. (2014). "Genetic structure of expanding wolf (Canis lupus) populations in Italy and Croatia, and the early steps of the recolonization of the Eastern Alps". Mammalian Biology - Zeitschrift für Säugetierkunde. 79 (2): 138–148. doi:10.1016/j.mambio.2013.10.002.
- Stronen, Astrid V.; Jä™Drzejewska, Bogumiå'a; Pertoldi, Cino; Demontis, Ditte; Randi, Ettore; Niedziaå'Kowska, Magdalena; Pilot, MaÅ'Gorzata; Sidorovich, Vadim E.; Dykyy, Ihor; Kusak, Josip; Tsingarska, Elena; Kojola, Ilpo; Karamanlidis, Alexandros A.; Ornicans, Aivars; Lobkov, Vladimir A.; Dumenko, Vitalii; Czarnomska, Sylwia D. (2013). "North-South Differentiation and a Region of High Diversity in European Wolves (Canis lupus)". PLOS ONE. 8 (10): e76454. doi:10.1371/journal.pone.0076454. PMC 3795770. PMID 24146871.
- Schweizer, Rena M.; Robinson, Jacqueline; Harrigan, Ryan; Silva, Pedro; Galverni, Marco; Musiani, Marco; Green, Richard E.; Novembre, John; Wayne, Robert K. (2016). "Targeted capture and resequencing of 1040 genes reveal environmentally driven functional variation in grey wolves". Molecular Ecology. 25 (1): 357–79. doi:10.1111/mec.13467. PMID 26562361. S2CID 17798894.
- O'Keefe, F. Robin; Meachen, Julie; Fet, Elizabeth V.; Brannick, Alexandria (2013). "Ecological determinants of clinal morphological variation in the cranium of the North American gray wolf". Journal of Mammalogy. 94 (6): 1223–1236. doi:10.1644/13-MAMM-A-069.
- Carmichael, L. E.; Krizan, J.; Nagy, J. A.; Fuglei, E.; Dumond, M.; Johnson, D.; Veitch, A.; Berteaux, D.; Strobeck, C. (2007). "Historical and ecological determinants of genetic structure in arctic canids". Molecular Ecology. 16 (16): 3466–83. doi:10.1111/j.1365-294X.2007.03381.x. PMID 17688546. S2CID 20760315.
- Carmichael, L. E.; Nagy, J. A.; Larter, N. C.; Strobeck, C. (2001). "Prey specialization may influence patterns of gene flow in wolves of the Canadian Northwest". Molecular Ecology. 10 (12): 2787–98. doi:10.1046/j.0962-1083.2001.01408.x. PMID 11903892. S2CID 29313917.
- Carmichael, L. E. (2006). Ecological Genetics of Northern Wolves and Arctic Foxes. Ph.D. Dissertation (PDF). University of Alberta.
- Lacombat, F.; Abbazzi, L.; Ferretti, M. P.; Martinez-Navarro, B.; Moulle, P.-E.; Palombo, M.-R.; Rook, L.; Turner, A.; Valli, A.M.-F. (2008). "New data on the early Villafranchian fauna from Vialette (Haute-Loire, France) based on the collection of the Crozatier Museum (Le Puy-en-Velay, Haute-Loire, France)". Quaternary International. 179 (1): 64–71. doi:10.1016/j.quaint.2007.09.005.
- Rook, L.; Torre, D. (1996). "The wolf-event in western Europe and the beginning of the Late Villafranchian". Neues Jahrbuch für Geologie und Paläontologie - Monatshefte. 1996 (8): 495–501. doi:10.1127/njgpm/1996/1996/495.
- Gliozzi, E.; Abbazzi, L.; Argenti, A.; Azzaroli, A.; Caloi, L.; Capasso Barbato, L.; di Stefano, G.; Esu, D.; Ficcarelli, G.; Girotti, O.; Kotsakis, T.; Masini, F.; Mazza, P.; Mezzabotta, C.; Palombo, M. R.; Petronio, C.; Rook, L.; Sala, B.; Sardella, R.; Zanalda, E.; Torre, D. (1997). "Biochronology of selected mammals, Molluscs and ostracodes from the Middle Pliocene to the Late Pleistocene in Italy. The state of the art" (PDF). Rivista Italiana di Paleontologia e Stratigrafia. 103 (3): 369–388. doi:10.13130/2039-4942/5299.
- Currant, A. P. (1984). "The mammalian remains". In Green, H. S. (ed.). Pontnewydd Cave: A Lower Palaeolithic Hominid Site in Wales: the First Report. Journal of Archaeological Science. 12. Amgueddfa Genedlaethol Cymru. pp. 177–181. doi:10.1016/0305-4403(85)90038-X. ISBN 978-0-7200-0282-9.
- Fox-Dobbs, Kena; Leonard, Jennifer A.; Koch, Paul L. (2008). "Pleistocene megafauna from eastern Beringia: Paleoecological and paleoenvironmental interpretations of stable carbon and nitrogen isotope and radiocarbon records". Palaeogeography, Palaeoclimatology, Palaeoecology. 261 (1–2): 30–46. doi:10.1016/j.palaeo.2007.12.011.
- Baryshnikov, Gennady F.; Mol, Dick; Tikhonov, Alexei N (2009). "Finding of the Late Pleistocene carnivores in Taimyr Peninsula (Russia, Siberia) with paleoecological context" (PDF). Russian Journal of Theriology. 8 (2): 107–113. doi:10.15298/rusjtheriol.08.2.04. Retrieved December 23, 2014.
- Ovodov, N. (2011). "A 33,000-year-old incipient dog from the Altai Mountains of Siberia: Evidence of the earliest domestication disrupted by the Last Glacial Maximum". PLOS ONE. 6 (7): e22821. Bibcode:2011PLoSO...622821O. doi:10.1371/journal.pone.0022821. PMC 3145761. PMID 21829526.
- Germonpré, Mietje; Lázničková-Galetová, Martina; Losey, Robert J.; Räikkönen, Jannikke; Sablin, Mikhail V. (2015). "Large canids at the Gravettian Předmostí site, the Czech Republic: The mandible". Quaternary International. 359–360: 261–279. doi:10.1016/j.quaint.2014.07.012.
- Germonpré, Mietje; Laznickova-Galetova, Martina; Sablin, Mikhail V. (2012). "Palaeolithic dog skulls at the Gravettian Predmosti site, the Czech Republic". Journal of Archaeological Science. 39: 184–202. doi:10.1016/j.jas.2011.09.022.
- Boudadi-Maligne, M. (2010). "Les Canis pleistocenes du sud de la France: approche biosystematique, evolutive et biochronologique. Ph.D. dissertation" (PDF). Archéologie et Préhistoire. Universitie Bordeaux (4126).
- Dimitrijević, V.; Vuković, S. (2015). "Was the Dog Locally Domesticated in the Danube Gorges? Morphometric Study of Dog Cranial Remains from Four Mesolithic-Early Neolithic Archaeological Sites by Comparison with Contemporary Wolves". International Journal of Osteoarchaeology. 25: 1–30. doi:10.1002/oa.2260.
- Germonpréa, Mietje; Sablinb, Mikhail V.; Stevensc, Rhiannon E.; Hedgesd, Robert E. M.; Hofreitere, Michael; Stillere, Mathias; Desprése, Viviane R. (2008). "Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: osteometry, ancient DNA and stable isotopes". Journal of Archaeological Science. 36 (2): 473–490. doi:10.1016/j.jas.2008.09.033.
- Larson G (2012). "Rethinking dog domestication by integrating genetics, archeology, and biogeography". PNAS. 109 (23): 8878–8883. Bibcode:2012PNAS..109.8878L. doi:10.1073/pnas.1203005109. PMC 3384140. PMID 22615366.
