KAT5
Histone acetyltransferase KAT5 is an enzyme that in humans is encoded by the KAT5 gene.[5][6] It is also commonly identified as TIP60.
The protein encoded by this gene belongs to the MYST family of histone acetyl transferases (HATs) and was originally isolated as an HIV-1 TAT-interactive protein. HATs play important roles in regulating chromatin remodeling, transcription and other nuclear processes by acetylating histone and nonhistone proteins. This protein is a histone acetylase that has a role in DNA repair and apoptosis and is thought to play an important role in signal transduction. Alternative splicing of this gene results in multiple transcript variants.[6]
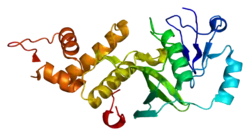
Structure
The structure of KAT5 includes an acetyl CoA binding domain and a zinc finger in the MYST domain, and a CHROMO domain.[7] Excess acetyl CoA is necessary for acetylation of histones. The zinc finger domain has been shown to aid in the acetylation process as well.[8] The CHROMO domain aids in KAT5 ability to bind chromatin, which is important for DNA repair.[9]
Function
KAT5 enzyme is known for acetylating histones in the nucleosome, which alters binding with DNA. Acetylation neutralizes the positive charge on histones, decreasing binding affinity of negatively charged DNA.[10] This in turn decreases steric hindrance of DNA and increases interaction of transcription factors and other proteins. Three key functions of KAT5 are its ability to regulate transcription, DNA repair, and apoptosis.
Transcription
Transcription factors such as E2F proteins and c-Myc can regulate the expression of proteins, particularly those involved with the cell cycle.[11][12] KAT5 acetylates histones on genes of these transcription factors, which promote their activity.
DNA repair
KAT5 is an important enzyme for repairing DNA and returning cellular function to normal through its regulation of ataxia telangiectasia mutant (ATM) protein kinase.[13] ATM protein kinase phosphorylates and therefore activates proteins involved in DNA repair. However, to be functional, ATM protein kinase must be acetylated by the KAT5 protein. Lack of KAT5 suppresses ATM protein kinase activity and reduces the ability of a cell to correct its DNA.
KAT5 also works later in the DNA repair process, as it serves as a cofactor for TRRAP.[14] TRRAP enhances DNA remodeling by binding to chromatin near broken double stranded DNA sequences. KAT5 aids this recognition.
Regulation
KAT5 catalytic activity is regulated by the phosphorylation of its histones during the G2/M phase of the cell cycle.[15] Phosphorylation of KAT5 serines 86 and 90 reduces its activity. Therefore, cancer cells with uncontrolled growth and improper G2/M checkpoints lack KAT5 regulation by cyclin dependent kinase (CDK) phosphorylation.
Clinical relevance
KAT5 has many clinically significant implications that make it a useful target for diagnostic or therapeutic approaches. Most notably, KAT5 helps to regulate cancers, HIV, and neurodegenerative diseases.[7]
Cancer
As mentioned above, KAT5 helps to repair DNA and upregualte tumor suppressors such as p53. Therefore, many cancers are marked by a reduction of KAT5 mRNA. KAT5 also is linked to metastasis and malignancy.[16]
- Colon cancer[17]
- Lung cancer[11]
- Breast cancer[18]
- Pancreatic[18]
- Gastric cancer[19]
- Metastatic melanoma[16]
Studies have also shown that KAT5 augmented the ability of chemotherapy to stop tumor growth, demonstrating its potential for use in combination therapy.[18]
However, KAT5 isn't always anti-cancer. It can enhance the activity of proteins for viruses that cause cancer such as human T-cell lymphotropic virus type-1 (HTLV), which may result in leukemia and lymphoma.[20] Additionally, KAT5 reacts with human papillomavirus (HPV), the virus responsible for cervical cancer.[21]
Other proteins that KAT5 promotes may lead to cancer as well. For example, overexpressed E2F1, a transcriptional factor, is implicated in melanoma progression.[22] More research needs to be performed to clearly elucidate the overall role KAT5 has in cancer.
Aging and Neurodegeneration
TIP60 regulates diverse cellular pathways including autophagy, DNA repair, neuronal survival, learning/memory, sleep/wake patterns, and protein turnover, all of which contribute to cellular homeostasis and organismal health so as to counteract aging and neurodegeneration.[24]
Interactions
HTATIP has been shown to interact with:
References
- GRCh38: Ensembl release 89: ENSG00000172977 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000024926 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G (Feb 1996). "Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator". Virology. 216 (2): 357–66. doi:10.1006/viro.1996.0071. PMID 8607265.
- "Entrez Gene: HTATIP HIV-1 Tat interacting protein, 60kDa".
- Mattera, L (2011). "HTATIP (HIV-1 Tat interacting protein, 60kDa)". Atlas of Genetics and Cytogenetics in Oncology and Haematology (3). doi:10.4267/2042/38522. hdl:2042/38522.
- Kim MY, Ann EJ, Kim JY, Mo JS, Park JH, Kim SY, Seo MS, Park HS (Sep 2007). "Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation". Molecular and Cellular Biology. 27 (18): 6506–19. doi:10.1128/MCB.01515-06. PMC 2099611. PMID 17636029.
- Koonin EV, Zhou S, Lucchesi JC (Nov 1995). "The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin". Nucleic Acids Research. 23 (21): 4229–33. doi:10.1093/nar/23.21.4229. PMC 307373. PMID 7501439.
- Lee, Frank. "Gene Transcription: Histone Acetylation, DNA Methylation and Epigenetics". Molecular Biology Web Book. Web Books Publishing.
- Van Den Broeck A, Nissou D, Brambilla E, Eymin B, Gazzeri S (Feb 2012). "Activation of a Tip60/E2F1/ERCC1 network in human lung adenocarcinoma cells exposed to cisplatin". Carcinogenesis. 33 (2): 320–5. doi:10.1093/carcin/bgr292. PMID 22159227.
- Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB (Dec 2004). "The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60". Molecular and Cellular Biology. 24 (24): 10826–34. doi:10.1128/MCB.24.24.10826-10834.2004. PMC 533976. PMID 15572685.
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD (Sep 2005). "A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM". Proceedings of the National Academy of Sciences of the United States of America. 102 (37): 13182–7. Bibcode:2005PNAS..10213182S. doi:10.1073/pnas.0504211102. PMC 1197271. PMID 16141325.
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z (Jan 2006). "Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks". Nature Cell Biology. 8 (1): 91–9. doi:10.1038/ncb1343. PMID 16341205. S2CID 25051471.
- Lemercier C, Legube G, Caron C, Louwagie M, Garin J, Trouche D, Khochbin S (Feb 2003). "Tip60 acetyltransferase activity is controlled by phosphorylation". The Journal of Biological Chemistry. 278 (7): 4713–8. doi:10.1074/jbc.M211811200. PMID 12468530. S2CID 8276821.
- Chen G, Cheng Y, Tang Y, Martinka M, Li G (Nov 2012). "Role of Tip60 in human melanoma cell migration, metastasis, and patient survival". The Journal of Investigative Dermatology. 132 (11): 2632–41. doi:10.1038/jid.2012.193. PMID 22673729.
- Chevillard-Briet M, Quaranta M, Grézy A, Mattera L, Courilleau C, Philippe M, Mercier P, Corpet D, Lough J, Ueda T, Fukunaga R, Trouche D, Escaffit F (Apr 2014). "Interplay between chromatin-modifying enzymes controls colon cancer progression through Wnt signaling". Human Molecular Genetics. 23 (8): 2120–31. doi:10.1093/hmg/ddt604. PMID 24287617.
- Ravichandran P, Ginsburg D (April 2015). "Tip60 Overexpression Exacerbates Chemotherapeutic Drug Treatment in Breast, Pancreatic, and Lung Cancer Cell Lines". The FASEB Journal. 29: Supplement 725.21. doi:10.1096/fasebj.29.1_supplement.725.21 (inactive 2021-01-14).CS1 maint: DOI inactive as of January 2021 (link)
- Sakuraba K, Yokomizo K, Shirahata A, Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Hibi K (Jan 2011). "TIP60 as a potential marker for the malignancy of gastric cancer". Anticancer Research. 31 (1): 77–9. PMID 21273583.
- Awasthi S, Sharma A, Wong K, Zhang J, Matlock EF, Rogers L, Motloch P, Takemoto S, Taguchi H, Cole MD, Lüscher B, Dittrich O, Tagami H, Nakatani Y, McGee M, Girard AM, Gaughan L, Robson CN, Monnat RJ, Harrod R (Jul 2005). "A human T-cell lymphotropic virus type 1 enhancer of Myc transforming potential stabilizes Myc-TIP60 transcriptional interactions". Molecular and Cellular Biology. 25 (14): 6178–98. doi:10.1128/MCB.25.14.6178-6198.2005. PMC 1168837. PMID 15988028.
- Hong S, Dutta A, Laimins LA (Apr 2015). "The acetyltransferase Tip60 is a critical regulator of the differentiation-dependent amplification of human papillomaviruses". Journal of Virology. 89 (8): 4668–75. doi:10.1128/JVI.03455-14. PMC 4442364. PMID 25673709.
- Alla V, Engelmann D, Niemetz A, Pahnke J, Schmidt A, Kunz M, Emmrich S, Steder M, Koczan D, Pützer BM (Jan 2010). "E2F1 in melanoma progression and metastasis". Journal of the National Cancer Institute. 102 (2): 127–33. doi:10.1093/jnci/djp458. PMID 20026813.
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G (Feb 1996). "Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator". Virology. 216 (2): 357–66. doi:10.1006/viro.1996.0071. PMID 8607265.
- Li, Zhiquan; Rasmussen, Lene Juel (2020-10-19). "TIP60 in Aging and Neurodegeneration". Ageing Research Reviews. 64: 101195. doi:10.1016/j.arr.2020.101195. ISSN 1568-1637. PMID 33091598. S2CID 224775578.
- Gaughan L, Logan IR, Cook S, Neal DE, Robson CN (Jul 2002). "Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor". The Journal of Biological Chemistry. 277 (29): 25904–13. doi:10.1074/jbc.M203423200. PMID 11994312. S2CID 9930504.
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, Scheidereit C, Leutz A (Jun 1999). "The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators". Oncogene. 18 (22): 3316–23. doi:10.1038/sj.onc.1202717. PMID 10362352. S2CID 2356435.
- Gavaravarapu S, Kamine J (Mar 2000). "Tip60 inhibits activation of CREB protein by protein kinase A". Biochemical and Biophysical Research Communications. 269 (3): 758–66. doi:10.1006/bbrc.2000.2358. PMID 10720489.
- Nordentoft I, Jørgensen P (Aug 2003). "The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor". The Biochemical Journal. 374 (Pt 1): 165–73. doi:10.1042/BJ20030087. PMC 1223570. PMID 12737628.
- Lee HJ, Chun M, Kandror KV (May 2001). "Tip60 and HDAC7 interact with the endothelin receptor a and may be involved in downstream signaling". The Journal of Biological Chemistry. 276 (20): 16597–600. doi:10.1074/jbc.C000909200. PMID 11262386. S2CID 38498534.
- Hejna J, Holtorf M, Hines J, Mathewson L, Hemphill A, Al-Dhalimy M, Olson SB, Moses RE (Apr 2008). "Tip60 is required for DNA interstrand cross-link repair in the Fanconi anemia pathway". The Journal of Biological Chemistry. 283 (15): 9844–51. doi:10.1074/jbc.M709076200. PMC 2398728. PMID 18263878.
- Xiao H, Chung J, Kao HY, Yang YC (Mar 2003). "Tip60 is a co-repressor for STAT3". The Journal of Biological Chemistry. 278 (13): 11197–204. doi:10.1074/jbc.M210816200. PMID 12551922. S2CID 6317335.
- Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D (Apr 2002). "Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation". The EMBO Journal. 21 (7): 1704–12. doi:10.1093/emboj/21.7.1704. PMC 125958. PMID 11927554.
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B (Jun 2003). "MYC recruits the TIP60 histone acetyltransferase complex to chromatin". EMBO Reports. 4 (6): 575–80. doi:10.1038/sj.embor.embor861. PMC 1319201. PMID 12776177.
- Sheridan AM, Force T, Yoon HJ, O'Leary E, Choukroun G, Taheri MR, Bonventre JV (Jul 2001). "PLIP, a novel splice variant of Tip60, interacts with group IV cytosolic phospholipase A(2), induces apoptosis, and potentiates prostaglandin production". Molecular and Cellular Biology. 21 (14): 4470–81. doi:10.1128/MCB.21.14.4470-4481.2001. PMC 87107. PMID 11416127.
- Bakshi, K., Ranjitha, B., Dubey, S. et al. Novel complex of HAT protein TIP60 and nuclear receptor PXR promotes cell migration and adhesion. Sci Rep 7, 3635 (2017). https://doi.org/10.1038/s41598-017-03783-w
Further reading
- Doyon Y, Côté J (Apr 2004). "The highly conserved and multifunctional NuA4 HAT complex". Current Opinion in Genetics & Development. 14 (2): 147–54. doi:10.1016/j.gde.2004.02.009. PMID 15196461.
- Sapountzi V, Logan IR, Robson CN (2006). "Cellular functions of TIP60". The International Journal of Biochemistry & Cell Biology. 38 (9): 1496–509. doi:10.1016/j.biocel.2006.03.003. PMID 16698308.
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Yamamoto T, Horikoshi M (Dec 1997). "Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60". The Journal of Biological Chemistry. 272 (49): 30595–8. doi:10.1074/jbc.272.49.30595. PMID 9388189. S2CID 21873080.
- Kimura A, Horikoshi M (Dec 1998). "Tip60 acetylates six lysines of a specific class in core histones in vitro". Genes to Cells. 3 (12): 789–800. doi:10.1046/j.1365-2443.1998.00229.x. PMID 10096020. S2CID 41070266.
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, Scheidereit C, Leutz A (Jun 1999). "The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators". Oncogene. 18 (22): 3316–23. doi:10.1038/sj.onc.1202717. PMID 10362352. S2CID 2356435.
- Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN (Jun 1999). "Tip60 is a nuclear hormone receptor coactivator". The Journal of Biological Chemistry. 274 (25): 17599–604. doi:10.1074/jbc.274.25.17599. PMID 10364196. S2CID 38058299.
- Creaven M, Hans F, Mutskov V, Col E, Caron C, Dimitrov S, Khochbin S (Jul 1999). "Control of the histone-acetyltransferase activity of Tip60 by the HIV-1 transactivator protein, Tat". Biochemistry. 38 (27): 8826–30. doi:10.1021/bi9907274. PMID 10393559.
- Sliva D, Zhu YX, Tsai S, Kamine J, Yang YC (Sep 1999). "Tip60 interacts with human interleukin-9 receptor alpha-chain". Biochemical and Biophysical Research Communications. 263 (1): 149–55. doi:10.1006/bbrc.1999.1083. PMID 10486269.
- Gavaravarapu S, Kamine J (Mar 2000). "Tip60 inhibits activation of CREB protein by protein kinase A". Biochemical and Biophysical Research Communications. 269 (3): 758–66. doi:10.1006/bbrc.2000.2358. PMID 10720489.
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG (Jul 2000). "Proteomic analysis of NMDA receptor-adhesion protein signaling complexes". Nature Neuroscience. 3 (7): 661–9. doi:10.1038/76615. hdl:1842/742. PMID 10862698. S2CID 14392630.
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y (Aug 2000). "Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis". Cell. 102 (4): 463–73. doi:10.1016/S0092-8674(00)00051-9. PMID 10966108. S2CID 18047169.
- Ran Q, Pereira-Smith OM (Nov 2000). "Identification of an alternatively spliced form of the Tat interactive protein (Tip60), Tip60(beta)". Gene. 258 (1–2): 141–6. doi:10.1016/S0378-1119(00)00410-8. PMID 11111051.
- Lee HJ, Chun M, Kandror KV (May 2001). "Tip60 and HDAC7 interact with the endothelin receptor a and may be involved in downstream signaling". The Journal of Biological Chemistry. 276 (20): 16597–600. doi:10.1074/jbc.C000909200. PMID 11262386. S2CID 38498534.
- Hlubek F, Löhberg C, Meiler J, Jung A, Kirchner T, Brabletz T (Apr 2001). "Tip60 is a cell-type-specific transcriptional regulator". Journal of Biochemistry. 129 (4): 635–41. doi:10.1093/oxfordjournals.jbchem.a002901. PMID 11275565.
- Sheridan AM, Force T, Yoon HJ, O'Leary E, Choukroun G, Taheri MR, Bonventre JV (Jul 2001). "PLIP, a novel splice variant of Tip60, interacts with group IV cytosolic phospholipase A(2), induces apoptosis, and potentiates prostaglandin production". Molecular and Cellular Biology. 21 (14): 4470–81. doi:10.1128/MCB.21.14.4470-4481.2001. PMC 87107. PMID 11416127.
- Cao X, Südhof TC (Jul 2001). "A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60". Science. 293 (5527): 115–20. doi:10.1126/science.1058783. PMID 11441186. S2CID 43920642.
- Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D (Apr 2002). "Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation". The EMBO Journal. 21 (7): 1704–12. doi:10.1093/emboj/21.7.1704. PMC 125958. PMID 11927554.