DNA damage (naturally occurring)
DNA damage is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of standard base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly.[1]
DNA damage and mutation have different biological consequences. While most DNA damages can undergo DNA repair, such repair is not 100% efficient. Un-repaired DNA damages accumulate in non-replicating cells, such as cells in the brains or muscles of adult mammals, and can cause aging.[2][3][4] (Also see DNA damage theory of aging.) In replicating cells, such as cells lining the colon, errors occur upon replication past damages in the template strand of DNA or during repair of DNA damages. These errors can give rise to mutations or epigenetic alterations.[5] Both of these types of alteration can be replicated and passed on to subsequent cell generations. These alterations can change gene function or regulation of gene expression and possibly contribute to progression to cancer.
Throughout the cell cycle there are various checkpoints to ensure the cell is in good condition to progress to mitosis. The three main checkpoints are at G1/s, G2/m, and at the spindle assembly checkpoint regulating progression through anaphase. G1 and G2 checkpoints involve scanning for damaged DNA.[6] During S phase the cell is more vulnerable to DNA damage than any other part of the cell cycle. G2 checkpoint checks for damaged DNA and DNA replication completeness. DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a base missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage.[7]
Types
Damage to DNA that occurs naturally can result from metabolic or hydrolytic processes. Metabolism releases compounds that damage DNA including reactive oxygen species, reactive nitrogen species, reactive carbonyl species, lipid peroxidation products and alkylating agents, among others, while hydrolysis cleaves chemical bonds in DNA.[8] Naturally occurring oxidative DNA damages arise at least 10,000 times per cell per day in humans and as much as 100,000 per cell per day in rats[9] as documented below.
Oxidative DNA damage can produce more than 20 types of altered bases[10][11] as well as single strand breaks.[12]
Other types of endogeneous DNA damages, given below with their frequencies of occurrence, include depurinations, depyrimidinations, double-strand breaks, O6-methylguanines and cytosine deamination.
DNA can be damaged via environmental factors as well. Environmental agents such as UV light, ionizing radiation, and genotoxic chemicals. Replication forks can be stalled due to damaged DNA and double strand breaks are also a form of DNA damage.[13]
Frequencies
The list below shows some frequencies with which new naturally occurring DNA damages arise per day, due to endogenous cellular processes.
- Oxidative damages
- Humans, per cell per day
- Rats, per cell per day
- Mice, per cell per day
- Depurinations
- Depyrimidinations
- Single-strand breaks
- Mammalian cells, per cell per day
- 55,200[23]
- Mammalian cells, per cell per day
- Double-strand breaks
- O6-methylguanines
- Mammalian cells, per cell per day
- 3,120[23]
- Mammalian cells, per cell per day
- Cytosine deamination
- Mammalian cells, per cell per day
- 192[23]
- Mammalian cells, per cell per day
Another important endogenous DNA damage is M1dG, short for (3-(2'-deoxy-beta-D-erythro-pentofuranosyl)-pyrimido[1,2-a]-purin-10(3H)-one). The excretion in urine (likely reflecting rate of occurrence) of M1dG may be as much as 1,000-fold lower than that of 8-oxodG.[26] However, a more important measure may be the steady-state level in DNA, reflecting both rate of occurrence and rate of DNA repair. The steady-state level of M1dG is higher than that of 8-oxodG.[27] This points out that some DNA damages produced at a low rate may be difficult to repair and remain in DNA at a high steady-state level. Both M1dG[28] and 8-oxodG[29] are mutagenic.
Steady-state levels
Steady-state levels of DNA damages represent the balance between formation and repair. More than 100 types of oxidative DNA damage have been characterized, and 8-oxodG constitutes about 5% of the steady state oxidative damages in DNA.[18] Helbock et al.[14] estimated that there were 24,000 steady state oxidative DNA adducts per cell in young rats and 66,000 adducts per cell in old rats. This reflects the accumulation of DNA damage with age. DNA damage accumulation with age is further described in DNA damage theory of aging.
Swenberg et al.[30] measured average amounts of selected steady state endogenous DNA damages in mammalian cells. The seven most common damages they evaluated are shown in Table 1.
| Endogenous lesions | Number per cell |
|---|---|
| Abasic sites | 30,000 |
| N7-(2-hydroxethyl)guanine (7HEG) | 3,000 |
| 8-hydroxyguanine | 2,400 |
| 7-(2-oxoethyl)guanine | 1,500 |
| Formaldehyde adducts | 960 |
| Acrolein-deoxyguanine | 120 |
| Malondialdehyde-deoxyguanine | 60 |
Evaluating steady-state damages in specific tissues of the rat, Nakamura and Swenberg[31] indicated that the number of abasic sites varied from about 50,000 per cell in liver, kidney and lung to about 200,000 per cell in the brain.
Biomolecular pathways
Proteins promoting endogenous DNA damage were identified in a 2019 paper as the DNA "damage-up" proteins (DDPs).[32] The DDP mechanisms fall into 3 clusters:
- reactive oxygen increase by transmembrane transporters,
- chromosome loss by replisome binding,
- replication stalling by transcription factors.[32]
The DDP human homologs are over-represented in known cancer drivers, and their RNAs in tumors predict heavy mutagenesis and a poor prognosis.[32]
Repair of damaged DNA
In the presence of DNA damage, the cell can either repair the damage or induce cell death if the damage is beyond repair.
Types
The seven main types of DNA repair and one pathway of damage tolerance, the lesions they address, and the accuracy of the repair (or tolerance) are shown in this table. For a brief description of the steps in repair see DNA repair mechanisms or see each individual pathway.
| Repair pathway | Lesions | Accuracy | Ref. |
|---|---|---|---|
| Base excision repair | corrects DNA damage from oxidation, deamination and alkylation, also single-strand breaks | accurate | [33][34] |
| Nucleotide excision repair | oxidative endogenous lesions such as cyclopurine, sunlight-induced thymine dimers (cyclobutane dimers and pyrimidine (6-4) pyrimidone photoproducts) | accurate | [35][36][37] |
| Homology-directed repair | double-strand breaks in the mid-S phase or mid-G2 phase of the cell cycle | accurate | [38] |
| Non-homologous end joining | double-strand breaks if cells are in the G0 phase. the G1 phase or the G2 phase of the cell cycle | somewhat inaccurate | [38] |
| Microhomology-mediated end joining or alt-End joining | double-strand breaks in the S phase of the cell cycle | always inaccurate | [38] |
| DNA mismatch repair | base substitution mismatches and insertion-deletion mismatches generated during DNA replication | accurate | [39] |
| Direct reversal (MGMT and AlkB) | 6-O-methylguanine is reversed to guanine by MGMT, some other methylated bases are demethylated by AlkB | accurate | [40] |
| Translesion synthesis | DNA damage tolerance process that allows the DNA replication machinery to replicate past DNA lesions | may be inaccurate | [41] |
Aging and cancer
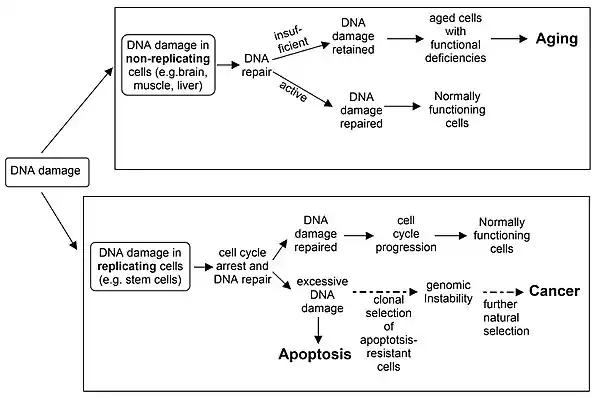
The schematic diagram indicates the roles of insufficient DNA repair in aging and cancer, and the role of apoptosis in cancer prevention. An excess of naturally occurring DNA damage, due to inherited deficiencies in particular DNA repair enzymes, can cause premature aging or increased risk for cancer (see DNA repair-deficiency disorder). On the other hand, the ability to trigger apoptosis in the presence of excess un-repaired DNA damage is critical for prevention of cancer.[42]
Apoptosis and cancer prevention
DNA repair proteins are often activated or induced when DNA has sustained damage. However, excessive DNA damage can initiate apoptosis (i.e., programmed cell death) if the level of DNA damage exceeds the repair capacity. Apoptosis can prevent cells with excess DNA damage from undergoing mutagenesis and progression to cancer.[43]
Inflammation is often caused by infection, such as with hepatitis B virus (HBV), hepatitis C virus (HCV) or Helicobacter pylori. Chronic inflammation is also a central characteristic of obesity.[44][45][46][47] Such inflammation causes oxidative DNA damage. This is due to the induction of reactive oxygen species (ROS) by various intracellular inflammatory mediators.[48][49][50] HBV and HCV infections, in particular, cause 10,000-fold and 100,000-fold increases in intracellular ROS production, respectively.[51] Inflammation-induced ROS that cause DNA damage can trigger apoptosis,[52][53] but may also cause cancer if repair and apoptotic processes are insufficiently protective.[45]
Bile acids, stored in the gall bladder, are released into the small intestine in response to fat in the diet. Higher levels of fat cause greater release.[54] Bile acids cause DNA damage, including oxidative DNA damage, double-strand DNA breaks, aneuploidy and chromosome breakage.[55] High-normal levels of the bile acid deoxycholic acid cause apoptosis in human colon cells,[56] but may also lead to colon cancer if repair and apoptotic defenses are insufficient.[57]
Apoptosis serves as a safeguard mechanism against tumorigenesis.[58] It prevents the increased mutagenesis that excess DNA damage could cause, upon replication.[59]
At least 17 DNA repair proteins, distributed among five DNA repair pathways, have a "dual role" in response to DNA damage. With moderate levels of DNA damage, these proteins initiate or contribute to DNA repair. However, when excessive levels of DNA damage are present, they trigger apoptosis.[43]
DNA damage response
The packaging of eukaryotic DNA into chromatin is a barrier to all DNA-based processes that require enzyme action. For most DNA repair processes, the chromatin must be remodeled. In eukaryotes, ATP-dependent chromatin remodeling complexes and histone-modifying enzymes are two factors that act to accomplish this remodeling process after DNA damage occurs.[60] Further DNA repair steps, involving multiple enzymes, usually follow. Some of the first responses to DNA damage, with their timing, are described below. More complete descriptions of the DNA repair pathways are presented in articles describing each pathway. At least 169 enzymes are involved in DNA repair pathways.[61]
Base excision repair
Oxidized bases in DNA are produced in cells treated with Hoechst dye followed by micro-irradiation with 405 nm light.[62] Such oxidized bases can be repaired by base excision repair.
When the 405 nm light is focused along a narrow line within the nucleus of a cell, about 2.5 seconds after irradiation, the chromatin remodeling enzyme Alc1 achieves half-maximum recruitment onto the irradiated micro-line.[63] The line of chromatin that was irradiated then relaxes, expanding side-to-side over the next 60 seconds.[63]
Within 6 seconds of the irradiation with 405 nm light, there is half-maximum recruitment of OGG1 to the irradiated line.[62] OGG1 is an enzyme that removes the oxidative DNA damage 8-oxo-dG from DNA. Removal of 8-oxo-dG, during base excision repair, occurs with a half-life of 11 minutes.[18]
Nucleotide excision repair
Ultraviolet (UV) light induces the formation of DNA damages including pyrimidine dimers (such as thymine dimers) and 6,4 photoproducts. These types of "bulky" damages are repaired by nucleotide excision repair.
After irradiation with UV light, DDB2, in a complex with DDB1, the ubiquitin ligase protein CUL4A and the RING finger protein ROC1, associates with sites of damage within chromatin. Half-maximum association occurs in 40 seconds.[64] PARP1 also associates within this period.[65] The PARP1 protein attaches to both DDB1 and DDB2 and then PARylates (creates a poly-ADP ribose chain) on DDB2 that attracts the DNA remodeling protein ALC1.[65] ALC1 relaxes chromatin at sites of UV damage to DNA. In addition, the ubiquitin E3 ligase complex DDB1-CUL4A carries out ubiquitination of the core histones H2A, H3, and H4, as well as the repair protein XPC, which has been attracted to the site of the DNA damage.[66] XPC, upon ubiquitination, is activated and initiates the nucleotide excision repair pathway. Somewhat later, at 30 minutes after UV damage, the INO80 chromatin remodeling complex is recruited to the site of the DNA damage, and this coincides with the binding of further nucleotide excision repair proteins, including ERCC1.[67]
Homologous recombinational repair
Double-strand breaks (DSBs) at specific sites can be induced by transfecting cells with a plasmid encoding I-SceI endonuclease (a homing endonuclease). Multiple DSBs can be induced by irradiating sensitized cells (labeled with 5'-bromo-2'-deoxyuridine and with Hoechst dye) with 780 nm light. These DSBs can be repaired by the accurate homologous recombinational repair or by the less accurate non-homologous end joining repair pathway. Here we describe the early steps in homologous recombinational repair (HRR).
After treating cells to introduce DSBs, the stress-activated protein kinase, c-Jun N-terminal kinase (JNK), phosphorylates SIRT6 on serine 10.[68] This post-translational modification facilitates the mobilization of SIRT6 to DNA damage sites with half-maximum recruitment in well under a second.[68] SIRT6 at the site is required for efficient recruitment of poly (ADP-ribose) polymerase 1 (PARP1) to a DNA break site and for efficient repair of DSBs.[68] PARP1 protein starts to appear at DSBs in less than a second, with half maximum accumulation within 1.6 seconds after the damage occurs.[69] This then allows half maximum recruitment of the DNA repair enzymes MRE11 within 13 seconds and NBS1 within 28 seconds.[69] MRE11 and NBS1 carry out early steps of the HRR pathway.
γH2AX, the phosphorylated form of H2AX is also involved in early steps of DSB repair. The histone variant H2AX constitutes about 10% of the H2A histones in human chromatin.[70] γH2AX (H2AX phosphorylated on serine 139) can be detected as soon as 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurs in one minute.[70] The extent of chromatin with phosphorylated γH2AX is about two million base pairs at the site of a DNA double-strand break.[70] γH2AX does not, itself, cause chromatin decondensation, but within 30 seconds of irradiation, RNF8 protein can be detected in association with γH2AX.[71] RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4,[72] a component of the nucleosome remodeling and deacetylase complex NuRD.
Pause for DNA repair
After rapid chromatin remodeling, cell cycle checkpoints may be activated to allow DNA repair to be completed before the cell cycle progresses. First, two kinases, ATM and ATR, are activated within 5 or 6 minutes after DNA is damaged. This is followed by phosphorylation of the cell cycle checkpoint protein Chk1, initiating its function, about 10 minutes after DNA is damaged.[73]
Role of oxidative damage to guanine in gene regulation
The DNA damage 8-oxo-dG does not occur randomly in the genome. In mouse embryonic fibroblasts, a 2 to 5-fold enrichment of 8-oxo-dG was found in genetic control regions, including promoters, 5'-untranslated regions and 3'-untranslated regions compared to 8-oxo-dG levels found in gene bodies and in intergenic regions.[74] In rat pulmonary artery endothelial cells, when 22,414 protein-coding genes were examined for locations of 8-oxo-dG, the majority of 8-oxo-dGs (when present) were found in promoter regions rather than within gene bodies.[75] Among hundreds of genes whose expression levels were affected by hypoxia, those with newly acquired promoter 8-oxo-dGs were upregulated, and those genes whose promoters lost 8-oxo-dGs were almost all downregulated.[75]
As reviewed by Wang et al.,[76] oxidized guanine appears to have multiple regulatory roles in gene expression. In particular, when oxidative stress produces 8-oxo-dG in the promoter of a gene, the oxidative stress may also inactivate OGG1, an enzyme that targets 8-oxo-dG and normally initiates repair of 8-oxo-dG damage. The inactive OGG1, which no longer excises 8-oxo-dG, nevertheless targets and complexes with 8-oxo-dG, and causes a sharp (~70o) bend in the DNA. This allows the assembly of a transcriptional initiation complex, up-regulating transcription of the associated gene.[76][77]
When 8-oxo-dG is formed in a guanine rich, potential G-quadruplex-forming sequence (PQS) in the coding strand of a promoter, active OGG1 excises the 8-oxo-dG and generates an apurinic/apyrimidinic site (AP site). The AP site enables melting of the duplex to unmask the PQS, adopting a G-quadruplex fold (G4 structure/motif) that has a regulatory role in transcription activation.[76][78]
When 8-oxo-dG is complexed with active OGG1 it may then recruit chromatin remodelers to modulate gene expression. Chromodomain helicase DNA-binding protein 4 (CHD4), a component of the (NuRD) complex, is recruited by OGG1 to oxidative DNA damage sites. CHD4 then attracts DNA and histone methylating enzymes that repress transcription of associated genes.[76]
Role of DNA damage in memory formation
Oxidation of guanine
Oxidation of guanine, particularly within CpG sites, may be especially important in learning and memory. Methylation of cytosines occurs at 60–90% of CpG sites depending on the tissue type.[79] In the mammalian brain, ~62% of CpGs are methylated.[79] Methylation of CpG sites tends to stably silence genes.[80] More than 500 of these CpG sites are de-methylated in neuron DNA during memory formation and memory consolidation in the hippocampus[81][82] and cingulate cortex[82] regions of the brain. As indicated below, the first step in de-methylation of methylated cytosine at a CpG site is oxidation of the guanine to form 8-oxo-dG.
Role of oxidized guanine in DNA de-methylation
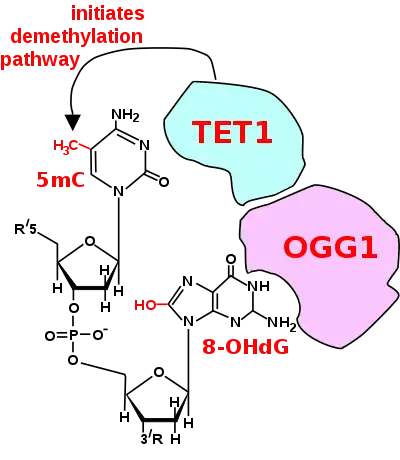
The figure in this section shows a CpG site where the cytosine is methylated to form 5-methylcytosine (5mC) and the guanine is oxidized to form 8-oxo-2'-deoxyguanosine (in the figure this is shown in the tautomeric form 8-OHdG). When this structure is formed, the base excision repair enzyme OGG1 targets 8-OHdG and binds to the lesion without immediate excision. OGG1, present at a 5mCp-8-OHdG site recruits TET1, and TET1 oxidizes the 5mC adjacent to the 8-OHdG. This initiates de-methylation of 5mC.[83] TET1 is a key enzyme involved in de-methylating 5mCpG. However, TET1 is only able to act on 5mCpG if the guanine was first oxidized to form 8-hydroxy-2'-deoxyguanosine (8-OHdG or its tautomer 8-oxo-dG), resulting in a 5mCp-8-OHdG dinucleotide (see figure in this section).[83] This initiates the de-methylation pathway on the methylated cytosine, finally resulting in an unmethylated cytosine (see DNA oxidation for further steps in forming unmethylated cytosine).
Altered protein expression in neurons, due to changes in methylation of DNA, (likely controlled by 8-oxo-dG-dependent de-methylation of CpG sites in gene promoters within neuron DNA) has been established as central to memory formation.[84]
Role of double-strand breaks in memory formation
Exposure of mice to physiological learning behaviors in vivo, such as being exposed to a new environment or activation of the primary visual cortex (V1) by exposing mice to visual stimuli, results in the formation of DNA double-strand breaks (DSBs) in the dentate gyrus (part of the hippocampus brain region).[85] Similarly, exposing mice to contextual fear conditioning, producing a long-term memory, also causes DSBs in the hippocampus within 15 minutes.[86] These neuronal activity-induced DSBs are restricted to only 21 loci in the neuron genome, and these loci are also enriched for the early response genes (including Fos, FosB, Npas4, Egr1, Nr4a1 and Nr4a3) in neuron activation.[86][87] These neural activity-induced DNA breaks are generated by a Type II topoisomerase.[86] An inhibitor of NHEJ DSB repair, ara-CTP, (likely inhibiting repair of such double-strand breaks), prevents long-term memory formation.[88]
Role of ATR and ATM
Most damage can be repaired without triggering the damage response system, however more complex damage activates ATR and ATM, key protein kinases in the damage response system.[89] DNA damage inhibits M-CDKs which are a key component of progression into Mitosis.
In all eukaryotic cells, ATR and ATM are protein kinases that detect DNA damage. They bind to DNA damaged sites and activate Chk1, Chk2, and, in animal cells, p53. Together, these proteins make up the DNA damage response system. Some DNA damage does not require the recruitment of ATR and ATM, it is only difficult and extensive damage that requires ATR and ATM. ATM and ATR are required for NHEJ, HR, ICL repair, and NER, as well as replication fork stability during unperturbed DNA replication and in response to replication blocks.[7]
ATR is recruited for different forms of damage such as nucleotide damage, stalled replication forks and double strand breaks. ATM is specifically for the damage response to double strand breaks. The MRN complex (composed of Mre11, Rad50, and Nbs1) form immediately at the site of double strand break. This MRN complex recruits ATM to the site of damage. ATR and ATM phosphorylate various proteins that contribute to the damage repair system. The binding of ATR and ATM to damage sites on DNA lead to the recruitment of Chk1 and Chk2. These protein kinases send damage signals to the cell cycle control system to delay the progression of the cell cycle.[13]
Chk1 and Chk2 functions
Chk1 leads to the production of DNA repair enzymes. Chk2 leads to reversible cell cycle arrest. Chk2, as well as ATR/ATM, can activate p53, which leads to permanent cell cycle arrest or apoptosis.
p53 role in DNA damage repair system
When there is too much damage, apoptosis is triggered in order to protect the organism from potentially harmful cells.7 p53, also known as a tumor suppressor gene, is a major regulatory protein in the DNA damage response system which binds directly to the promoters of its target genes. p53 acts primarily at the G1 checkpoint (controlling the G1 to S transition), where it blocks cell cycle progression.[6] Activation of p53 can trigger cell death or permanent cell cycle arrest. p53 can also activate certain repair pathways such was NER.[89]
Regulation of p53
In the absence of DNA damage, p53 is regulated by Mdm2 and constantly degraded. When there is DNA damage, Mdm2 is phosphorylated, most likely caused by ATM. The phosphorylation of Mdm2 leads to a reduction in the activity of Mdm2, thus preventing the degradation of p53. Normal, undamaged cell, usually has low levels of p53 while cells under stress and DNA damage, will have high levels of p53.[13]
p53 serves as transcription factor for bax and p21
p53 serves as a transcription factors for both bax, a proapoptotic protein as well as p21, a CDK inhibitor. CDK Inhibitors result in cell cycle arrest. Arresting the cell provides the cell time to repair the damage, and if the damage is irreparable, p53 recruits bax to trigger apoptosis.[89]
DDR and p53 role in cancer
p53 is a major key player in the growth of cancerous cells. Damaged DNA cells with mutated p53 are at a higher risk of becoming cancerous. Common chemotherapy treatments are genotoxic. These treatments are ineffective in cancer tumor that have mutated p53 since they do not have a functioning p53 to either arrest or kill the damaged cell.
A major problem for life
One indication that DNA damages are a major problem for life is that DNA repair processes, to cope with DNA damages, have been found in all cellular organisms in which DNA repair has been investigated. For example, in bacteria, a regulatory network aimed at repairing DNA damages (called the SOS response in Escherichia coli) has been found in many bacterial species. E. coli RecA, a key enzyme in the SOS response pathway, is the defining member of a ubiquitous class of DNA strand-exchange proteins that are essential for homologous recombination, a pathway that maintains genomic integrity by repairing broken DNA.[90] Genes homologous to RecA and to other central genes in the SOS response pathway are found in almost all the bacterial genomes sequenced to date, covering a large number of phyla, suggesting both an ancient origin and a widespread occurrence of recombinational repair of DNA damage.[91] Eukaryotic recombinases that are homologues of RecA are also widespread in eukaryotic organisms. For example, in fission yeast and humans, RecA homologues promote duplex-duplex DNA-strand exchange needed for repair of many types of DNA lesions.[92][93]
Another indication that DNA damages are a major problem for life is that cells make large investments in DNA repair processes. As pointed out by Hoeijmakers,[3] repairing just one double-strand break could require more than 10,000 ATP molecules, as used in signaling the presence of the damage, the generation of repair foci, and the formation (in humans) of the RAD51 nucleofilament (an intermediate in homologous recombinational repair). (RAD51 is a homologue of bacterial RecA.) If the structural modification occurs during the G1 phase of DNA replication, the G1-S checkpoint arrests or postpones the furtherance of the cell cycle before the product enters the S phase.[1]
Consequences
Differentiated somatic cells of adult mammals generally replicate infrequently or not at all. Such cells, including, for example, brain neurons and muscle myocytes, have little or no cell turnover. Non-replicating cells do not generally generate mutations due to DNA damage-induced errors of replication. These non-replicating cells do not commonly give rise to cancer, but they do accumulate DNA damages with time that likely contribute to aging (). In a non-replicating cell, a single-strand break or other type of damage in the transcribed strand of DNA can block RNA polymerase II-catalysed transcription.[94] This would interfere with the synthesis of the protein coded for by the gene in which the blockage occurred.
Brasnjevic et al.[95] summarized the evidence showing that single-strand breaks accumulate with age in the brain (though accumulation differed in different regions of the brain) and that single-strand breaks are the most frequent steady-state DNA damages in the brain. As discussed above, these accumulated single-strand breaks would be expected to block transcription of genes. Consistent with this, as reviewed by Hetman et al.,[96] 182 genes were identified and shown to have reduced transcription in the brains of individuals older than 72 years, compared to transcription in the brains of those less than 43 years old. When 40 particular proteins were evaluated in a muscle of rats, the majority of the proteins showed significant decreases during aging from 18 months (mature rat) to 30 months (aged rat) of age.[97]
Another type of DNA damage, the double-strand break, was shown to cause cell death (loss of cells) through apoptosis.[98] This type of DNA damage would not accumulate with age, since once a cell was lost through apoptosis, its double-strand damage would be lost with it. Thus, damaged DNA segments undermine the DNA replication machinery because these altered sequences of DNA cannot be utilized as true templates to produce copies of one's genetic material.[1]
RAD genes and the cell cycle response to DNA damage in Saccharomyces cerevisiae
When DNA is damaged, the cell responds in various ways to fix the damage and minimize the effects on the cell. One such response, specifically in eukaryotic cells, is to delay cell division—the cell becomes arrested for some time in the G2 phase before progressing through the rest of the cell cycle. Various studies have been conducted to elucidate the purpose of this G2 arrest that is induced by DNA damage. Researchers have found that cells that are prematurely forced out of the delay have lower cell viability and higher rates of damaged chromosomes compared with cells that are able to undergo a full G2 arrest, suggesting that the purpose of the delay is to give the cell time to repair damaged chromosomes before continuing with the cell cycle.[99] This ensures the proper functioning of mitosis.
Various species of animals exhibit similar mechanisms of cellular delay in response to DNA damage, which can be caused by exposure to x-irradiation. The budding yeast Saccharomyces cerevisiae has specifically been studied because progression through the cell cycle can be followed via nuclear morphology with ease. By studying Saccharomyces cerevisiae, researchers have been able to learn more about radiation-sensitive (RAD) genes, and the effect that RAD mutations may have on the typical cellular DNA damaged-induced delay response. Specifically, the RAD9 gene plays a crucial role in detecting DNA damage and arresting the cell in G2 until the damage is repaired.
Through extensive experiments, researchers have been able to illuminate the role that the RAD genes play in delaying cell division in response to DNA damage. When wild-type, growing cells are exposed to various levels of x-irradiation over a given time frame, and then analyzed with a microcolony assay, differences in the cell cycle response can be observed based on which genes are mutated in the cells. For instance, while unirradiated cells will progress normally through the cell cycle, cells that are exposed to x-irradiation either permanently arrest (become inviable) or delay in the G2 phase before continuing to divide in mitosis, further corroborating the idea that the G2 delay is crucial for DNA repair. However, rad strains, which are deficient in DNA repair, exhibit a markedly different response. For instance, rad52 cells, which cannot repair double-stranded DNA breaks, tend to permanently arrest in G2 when exposed to even very low levels of x-irradiation, and rarely end up progressing through the later stages of the cell cycle. This is because the cells cannot repair DNA damage and thus do not enter mitosis. Various other rad mutants exhibit similar responses when exposed to x-irradiation.
However, the rad9 strain exhibits an entirely different effect. These cells fail to delay in the G2 phase when exposed to x-irradiation, and end up progressing through the cell cycle unperturbed, before dying. This suggests that the RAD9 gene, unlike the other RAD genes, plays a crucial role in initiating G2 arrest. To further investigate these findings, the cell cycles of double mutant strains have been analyzed. A mutant rad52 rad9 strain—which is both defective in DNA repair and G2 arrest—fails to undergo cell cycle arrest when exposed to x-irradiation. This suggests that even if DNA damage cannot be repaired, if RAD9 is not present, the cell cycle will not delay. Thus, unrepaired DNA damage is the signal that tells RAD9 to halt division and arrest the cell cycle in G2. Furthermore, there is a dose-dependent response; as the levels of x-irradiation—and subsequent DNA damage—increase, more cells, regardless of the mutations they have, become arrested in G2.
Another, and perhaps more helpful way to visualize this effect is to look at photomicroscopy slides. Initially, slides of RAD+ and rad9 haploid cells in the exponential phase of growth show simple, single cells, that are indistinguishable from each other. However, the slides look much different after being exposed to x-irradiation for 10 hours. The RAD+ slides now show RAD+ cells existing primarily as two-budded microcolonies, suggesting that cell division has been arrested. In contrast, the rad9 slides show the rad9 cells existing primarily as 3 to 8 budded colonies, and they appear smaller than the RAD+ cells. This is further evidence that the mutant RAD cells continued to divide and are deficient in G2 arrest.
However, there is evidence that although the RAD9 gene is necessary to induce G2 arrest in response to DNA damage, giving the cell time to repair the damage, it does not actually play a direct role in repairing DNA. When rad9 cells are artificially arrested in G2 with MBC, a microtubule poison that prevents cellular division, and then treated with x-irradiation, the cells are able to repair their DNA and eventually progress through the cell cycle, dividing into viable cells. Thus, the RAD9 gene plays no role in actually repairing damaged DNA—it simply senses damaged DNA and responds by delaying cell division. The delay, then, is mediated by a control mechanism, rather than the physical damaged DNA.[100]
On the other hand, it is possible that there are backup mechanisms that fill the role of RAD9 when it is not present. In fact, some studies have found that RAD9 does indeed play a critical role in DNA repair. In one study, rad9 mutant and normal cells in the exponential phase of growth were exposed to UV-irradiation and synchronized in specific phases of the cell cycle. After being incubated to permit DNA repair, the extent of pyrimidine dimerization (which is indicative of DNA damage) was assessed using sensitive primer extension techniques. It was found that the removal of DNA photolesions was much less efficient in rad9 mutant cells than normal cells, providing evidence that RAD9 is involved in DNA repair. Thus, the role of RAD9 in repairing DNA damage remains unclear.[101]
Regardless, it is clear that RAD9 is necessary to sense DNA damage and halt cell division. RAD9 has been suggested to possess 3’ to 5’ exonuclease activity, which is perhaps why it plays a role in detecting DNA damage. When DNA is damaged, it is hypothesized that RAD9 forms a complex with RAD1 and HUS1, and this complex is recruited to sites of DNA damage. It is in this way that RAD9 is able to exert its effects.
Although the function of RAD9 has primarily been studied in the budding yeast Saccharomyces cerevisiae, many of the cell cycle control mechanisms are similar between species. Thus, we can conclude that RAD9 likely plays a critical role in the DNA damage response in humans as well.
See also
References
- Köhler K, Ferreira P, Pfander B, Boos D (2016). The Initiation of DNA Replication in Eukaryotes. Springer, Cham. pp. 443–460. doi:10.1007/978-3-319-24696-3_22. ISBN 9783319246949.
- Bernstein H, Payne CM, Bernstein C, Garewal H, Dvorak K (2008). Cancer and aging as consequences of un-repaired DNA damage. In: New Research on DNA Damages (Editors: Honoka Kimura and Aoi Suzuki) Nova Science Publishers, Inc., New York, Chapter 1, pp. 1–47. open access, but read only https://www.novapublishers.com/catalog/product_info.php?products_id=43247 Archived 2014-10-25 at the Wayback Machine ISBN 978-1604565812
- Hoeijmakers JH (October 2009). "DNA damage, aging, and cancer". The New England Journal of Medicine. 361 (15): 1475–85. doi:10.1056/NEJMra0804615. PMID 19812404.
- Freitas AA, de Magalhães JP (2011). "A review and appraisal of the DNA damage theory of ageing". Mutation Research. 728 (1–2): 12–22. doi:10.1016/j.mrrev.2011.05.001. PMID 21600302.
- O'Hagan HM, Mohammad HP, Baylin SB (August 2008). "Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island". PLOS Genetics. 4 (8): e1000155. doi:10.1371/journal.pgen.1000155. PMC 2491723. PMID 18704159.
- "Khan Academy". Khan Academy. Retrieved 2017-12-15.
- Ciccia A, Elledge SJ (October 2010). "The DNA damage response: making it safe to play with knives". Molecular Cell. 40 (2): 179–204. doi:10.1016/j.molcel.2010.09.019. PMC 2988877. PMID 20965415.
- De Bont R, van Larebeke N (May 2004). "Endogenous DNA damage in humans: a review of quantitative data". Mutagenesis. 19 (3): 169–85. doi:10.1093/mutage/geh025. PMID 15123782.
- Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):7915-22. doi: 10.1073/pnas.90.17.7915. PMID: 8367443; PMCID: PMC47258.
- Yu Y, Cui Y, Niedernhofer LJ, Wang Y (December 2016). "Occurrence, Biological Consequences, and Human Health Relevance of Oxidative Stress-Induced DNA Damage". Chemical Research in Toxicology. 29 (12): 2008–2039. doi:10.1021/acs.chemrestox.6b00265. PMC 5614522. PMID 27989142.
- Dizdaroglu M, Coskun E, Jaruga P (May 2015). "Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques". Free Radical Research. 49 (5): 525–48. doi:10.3109/10715762.2015.1014814. PMID 25812590. S2CID 31852987.
- Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, et al. (September 2004). "In situ analysis of repair processes for oxidative DNA damage in mammalian cells". Proceedings of the National Academy of Sciences of the United States of America. 101 (38): 13738–43. Bibcode:2004PNAS..10113738L. doi:10.1073/pnas.0406048101. PMC 518826. PMID 15365186.
- Morgan, David (2006). Cell Cycle: Principles of Control. London: New Science Press.
- Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN (January 1998). "DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine". Proceedings of the National Academy of Sciences of the United States of America. 95 (1): 288–93. Bibcode:1998PNAS...95..288H. doi:10.1073/pnas.95.1.288. PMC 18204. PMID 9419368.
- Foksinski M, Rozalski R, Guz J, Ruszkowska B, Sztukowska P, Piwowarski M, et al. (November 2004). "Urinary excretion of DNA repair products correlates with metabolic rates as well as with maximum life spans of different mammalian species". Free Radical Biology & Medicine. 37 (9): 1449–54. doi:10.1016/j.freeradbiomed.2004.07.014. PMID 15454284.
- Tudek B, Winczura A, Janik J, Siomek A, Foksinski M, Oliński R (May 2010). "Involvement of oxidatively damaged DNA and repair in cancer development and aging". American Journal of Translational Research. 2 (3): 254–84. PMC 2892402. PMID 20589166.
- Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN (June 1990). "Oxidative damage to DNA during aging: 8-hydroxy-2'-deoxyguanosine in rat organ DNA and urine". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4533–7. Bibcode:1990PNAS...87.4533F. doi:10.1073/pnas.87.12.4533. PMC 54150. PMID 2352934.
- Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, et al. (May 2001). "A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA". Nucleic Acids Research. 29 (10): 2117–26. doi:10.1093/nar/29.10.2117. PMC 55450. PMID 11353081.
- Lindahl T, Nyberg B (September 1972). "Rate of depurination of native deoxyribonucleic acid". Biochemistry. 11 (19): 3610–8. doi:10.1021/bi00769a018. PMID 4626532.
- Lindahl T (April 1993). "Instability and decay of the primary structure of DNA". Nature. 362 (6422): 709–15. Bibcode:1993Natur.362..709L. doi:10.1038/362709a0. PMID 8469282. S2CID 4283694.
- Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA (January 1998). "Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions". Cancer Research. 58 (2): 222–5. PMID 9443396.
- Lindahl T. (1977) DNA repair enzymes acting on spontaneous lesions in DNA. In: Nichols WW and Murphy DG (eds.) DNA Repair Processes. Symposia Specialists, Miami p225-240. ISBN 088372099X ISBN 978-0883720998
- Tice, R.R., and Setlow, R.B. (1985) DNA repair and replication in aging organisms and cells. In: Finch EE and Schneider EL (eds.) Handbook of the Biology of Aging. Van Nostrand Reinhold, New York. Pages 173–224. ISBN 0442225296 ISBN 978-0442225292
- Haber JE (July 1999). "DNA recombination: the replication connection". Trends in Biochemical Sciences. 24 (7): 271–5. doi:10.1016/s0968-0004(99)01413-9. PMID 10390616.
- Vilenchik MM, Knudson AG (October 2003). "Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer". Proceedings of the National Academy of Sciences of the United States of America. 100 (22): 12871–6. Bibcode:2003PNAS..10012871V. doi:10.1073/pnas.2135498100. PMC 240711. PMID 14566050.
- Chan SW, Dedon PC (December 2010). "The biological and metabolic fates of endogenous DNA damage products". Journal of Nucleic Acids. 2010: 929047. doi:10.4061/2010/929047. PMC 3010698. PMID 21209721.
- Kadlubar FF, Anderson KE, Häussermann S, Lang NP, Barone GW, Thompson PA, et al. (September 1998). "Comparison of DNA adduct levels associated with oxidative stress in human pancreas". Mutation Research. 405 (2): 125–33. doi:10.1016/s0027-5107(98)00129-8. PMID 9748537.
- VanderVeen LA, Hashim MF, Shyr Y, Marnett LJ (November 2003). "Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde". Proceedings of the National Academy of Sciences of the United States of America. 100 (24): 14247–52. Bibcode:2003PNAS..10014247V. doi:10.1073/pnas.2332176100. PMC 283577. PMID 14603032.
- Tan X, Grollman AP, Shibutani S (December 1999). "Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2'-deoxyadenosine and 8-oxo-7,8-dihydro-2'-deoxyguanosine DNA lesions in mammalian cells". Carcinogenesis. 20 (12): 2287–92. doi:10.1093/carcin/20.12.2287. PMID 10590221.
- Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB (March 2011). "Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment". Toxicological Sciences. 120 Suppl 1 (Suppl 1): S130-45. doi:10.1093/toxsci/kfq371. PMC 3043087. PMID 21163908.
- Nakamura J, Swenberg JA (June 1999). "Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues". Cancer Research. 59 (11): 2522–6. PMID 10363965.
- Xia J, Chiu LY, Nehring RB, Bravo Núñez MA, Mei Q, Perez M, et al. (January 2019). "Bacteria-to-Human Protein Networks Reveal Origins of Endogenous DNA Damage". Cell. 176 (1–2): 127–143.e24. doi:10.1016/j.cell.2018.12.008. PMC 6344048. PMID 30633903.
- Krokan HE, Bjørås M (April 2013). "Base excision repair". Cold Spring Harbor Perspectives in Biology. 5 (4): a012583. doi:10.1101/cshperspect.a012583. PMC 3683898. PMID 23545420.
- del Rivero J, Kohn EC (April 2017). "PARP Inhibitors: The Cornerstone of DNA Repair-Targeted Therapies". Oncology. 31 (4): 265–73. PMID 28412778.
- Schärer OD (October 2013). "Nucleotide excision repair in eukaryotes". Cold Spring Harbor Perspectives in Biology. 5 (10): a012609. doi:10.1101/cshperspect.a012609. PMC 3783044. PMID 24086042.
- de Boer J, Hoeijmakers JH (March 2000). "Nucleotide excision repair and human syndromes". Carcinogenesis. 21 (3): 453–60. doi:10.1093/carcin/21.3.453. PMID 10688865.
- Satoh MS, Jones CJ, Wood RD, Lindahl T (July 1993). "DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical-induced base lesions". Proceedings of the National Academy of Sciences of the United States of America. 90 (13): 6335–9. Bibcode:1993PNAS...90.6335S. doi:10.1073/pnas.90.13.6335. PMC 46923. PMID 8327515.
- Ceccaldi R, Rondinelli B, D'Andrea AD (January 2016). "Repair Pathway Choices and Consequences at the Double-Strand Break". Trends in Cell Biology. 26 (1): 52–64. doi:10.1016/j.tcb.2015.07.009. PMC 4862604. PMID 26437586.
- Kunkel TA, Erie DA (2005). "DNA mismatch repair". Annual Review of Biochemistry. 74: 681–710. doi:10.1146/annurev.biochem.74.082803.133243. PMID 15952900.
- Yi C, He C (January 2013). "DNA repair by reversal of DNA damage". Cold Spring Harbor Perspectives in Biology. 5 (1): a012575. doi:10.1101/cshperspect.a012575. PMC 3579392. PMID 23284047.
- Lehmann AR (February 2005). "Replication of damaged DNA by translesion synthesis in human cells". FEBS Letters. 579 (4): 873–6. doi:10.1016/j.febslet.2004.11.029. PMID 15680966. S2CID 38747288.
- Nowsheen S, Yang ES (October 2012). "The intersection between DNA damage response and cell death pathways". Experimental Oncology. 34 (3): 243–54. PMC 3754840. PMID 23070009.
- Bernstein C, Bernstein H, Payne CM, Garewal H (June 2002). "DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis". Mutation Research. 511 (2): 145–78. doi:10.1016/s1383-5742(02)00009-1. PMID 12052432.
- Deng T, Lyon CJ, Bergin S, Caligiuri MA, Hsueh WA (May 2016). "Obesity, Inflammation, and Cancer". Annual Review of Pathology. 11: 421–49. doi:10.1146/annurev-pathol-012615-044359. PMID 27193454.
- Iyengar NM, Gucalp A, Dannenberg AJ, Hudis CA (December 2016). "Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation". Journal of Clinical Oncology. 34 (35): 4270–4276. doi:10.1200/JCO.2016.67.4283. PMC 5562428. PMID 27903155.
- Ramos-Nino ME (2013). "The role of chronic inflammation in obesity-associated cancers". ISRN Oncology. 2013: 697521. doi:10.1155/2013/697521. PMC 3683483. PMID 23819063.
- "Obesity and Cancer". 2017.
- Coussens LM, Werb Z (2002). "Inflammation and cancer". Nature. 420 (6917): 860–7. Bibcode:2002Natur.420..860C. doi:10.1038/nature01322. PMC 2803035. PMID 12490959.
- Chiba T, Marusawa H, Ushijima T (September 2012). "Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation". Gastroenterology. 143 (3): 550–563. doi:10.1053/j.gastro.2012.07.009. hdl:2433/160134. PMID 22796521.
- Shacter E, Weitzman SA (February 2002). "Chronic inflammation and cancer". Oncology. 16 (2): 217–26, 229, discussion 230–2. PMID 11866137.
- Valgimigli M, Valgimigli L, Trerè D, Gaiani S, Pedulli GF, Gramantieri L, Bolondi L (September 2002). "Oxidative stress EPR measurement in human liver by radical-probe technique. Correlation with etiology, histology and cell proliferation". Free Radical Research. 36 (9): 939–48. doi:10.1080/107156021000006653. PMID 12448819. S2CID 12061790.
- Hardbower DM, de Sablet T, Chaturvedi R, Wilson KT (2013). "Chronic inflammation and oxidative stress: the smoking gun for Helicobacter pylori-induced gastric cancer?". Gut Microbes. 4 (6): 475–81. doi:10.4161/gmic.25583. PMC 3928159. PMID 23811829.
- Ernst P (March 1999). "Review article: the role of inflammation in the pathogenesis of gastric cancer". Alimentary Pharmacology & Therapeutics. 13 Suppl 1: 13–8. doi:10.1046/j.1365-2036.1999.00003.x. PMID 10209682. S2CID 38496014.
- Marciani L, Cox EF, Hoad CL, Totman JJ, Costigan C, Singh G, et al. (November 2013). "Effects of various food ingredients on gall bladder emptying". European Journal of Clinical Nutrition. 67 (11): 1182–7. doi:10.1038/ejcn.2013.168. PMC 3898429. PMID 24045793.
- Payne CM, Bernstein C, Dvorak K, Bernstein H (2008). "Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis". Clinical and Experimental Gastroenterology. 1: 19–47. doi:10.2147/ceg.s4343. PMC 3108627. PMID 21677822.
- Bernstein C, Bernstein H, Garewal H, Dinning P, Jabi R, Sampliner RE, et al. (May 1999). "A bile acid-induced apoptosis assay for colon cancer risk and associated quality control studies". Cancer Research. 59 (10): 2353–7. PMID 10344743.
- Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, Bernstein H (August 2011). "Carcinogenicity of deoxycholate, a secondary bile acid". Archives of Toxicology. 85 (8): 863–71. doi:10.1007/s00204-011-0648-7. PMC 3149672. PMID 21267546.
- Zhang L, Yu J (December 2013). "Role of apoptosis in colon cancer biology, therapy, and prevention". Current Colorectal Cancer Reports. 9 (4): 331–340. doi:10.1007/s11888-013-0188-z. PMC 3836193. PMID 24273467.
- Williams GT, Critchlow MR, Hedge VL, O'Hare KB (December 1998). "Molecular failure of apoptosis: inappropriate cell survival and mutagenesis?". Toxicology Letters. 102–103: 485–9. doi:10.1016/s0378-4274(98)00343-9. PMID 10022300.
- Liu B, Yip RK, Zhou Z (November 2012). "Chromatin remodeling, DNA damage repair and aging". Current Genomics. 13 (7): 533–47. doi:10.2174/138920212803251373. PMC 3468886. PMID 23633913.
- "Human DNA repair genes".
- Abdou I, Poirier GG, Hendzel MJ, Weinfeld M (January 2015). "DNA ligase III acts as a DNA strand break sensor in the cellular orchestration of DNA strand break repair". Nucleic Acids Research. 43 (2): 875–92. doi:10.1093/nar/gku1307. PMC 4333375. PMID 25539916.
- Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, et al. (December 2016). "The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage". Molecular Biology of the Cell. 27 (24): 3791–3799. doi:10.1091/mbc.E16-05-0269. PMC 5170603. PMID 27733626.
- Luijsterburg MS, Goedhart J, Moser J, Kool H, Geverts B, Houtsmuller AB, et al. (August 2007). "Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC". Journal of Cell Science. 120 (Pt 15): 2706–16. doi:10.1242/jcs.008367. PMID 17635991.
- Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, et al. (October 2012). "PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1". The Journal of Cell Biology. 199 (2): 235–49. doi:10.1083/jcb.201112132. PMC 3471223. PMID 23045548.
- Yeh JI, Levine AS, Du S, Chinte U, Ghodke H, Wang H, et al. (October 2012). "Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair". Proceedings of the National Academy of Sciences of the United States of America. 109 (41): E2737-46. doi:10.1073/pnas.1110067109. PMC 3478663. PMID 22822215.
- Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L (October 2010). "INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway". Proceedings of the National Academy of Sciences of the United States of America. 107 (40): 17274–9. Bibcode:2010PNAS..10717274J. doi:10.1073/pnas.1008388107. PMC 2951448. PMID 20855601.
- Van Meter M, Simon M, Tombline G, May A, Morello TD, Hubbard BP, et al. (September 2016). "JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks". Cell Reports. 16 (10): 2641–2650. doi:10.1016/j.celrep.2016.08.006. PMC 5089070. PMID 27568560.
- Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, Poirier GG (January 2008). "PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites". The Journal of Biological Chemistry. 283 (2): 1197–208. doi:10.1074/jbc.M706734200. PMID 18025084.
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (March 1998). "DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139". The Journal of Biological Chemistry. 273 (10): 5858–68. doi:10.1074/jbc.273.10.5858. PMID 9488723.
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (November 2007). "RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins". Cell. 131 (5): 887–900. doi:10.1016/j.cell.2007.09.040. PMID 18001824. S2CID 14232192.
- Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, et al. (May 2012). "A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure". The EMBO Journal. 31 (11): 2511–27. doi:10.1038/emboj.2012.104. PMC 3365417. PMID 22531782.
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (January 2006). "ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks". Nature Cell Biology. 8 (1): 37–45. doi:10.1038/ncb1337. PMID 16327781. S2CID 9797133.
- Ding Y, Fleming AM, Burrows CJ (February 2017). "Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-dihydroguanine by OG-Seq". Journal of the American Chemical Society. 139 (7): 2569–2572. doi:10.1021/jacs.6b12604. PMC 5440228. PMID 28150947.
- Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, et al. (December 2015). "An oxidative DNA "damage" and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression". American Journal of Physiology. Lung Cellular and Molecular Physiology. 309 (11): L1367-75. doi:10.1152/ajplung.00236.2015. PMC 4669343. PMID 26432868.
- Wang R, Hao W, Pan L, Boldogh I, Ba X (October 2018). "The roles of base excision repair enzyme OGG1 in gene expression". Cellular and Molecular Life Sciences. 75 (20): 3741–3750. doi:10.1007/s00018-018-2887-8. PMC 6154017. PMID 30043138.
- Seifermann M, Epe B (June 2017). "Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark?". Free Radical Biology & Medicine. 107: 258–265. doi:10.1016/j.freeradbiomed.2016.11.018. PMID 27871818.
- Fleming AM, Burrows CJ (August 2017). "8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis". DNA Repair. 56: 75–83. doi:10.1016/j.dnarep.2017.06.009. PMC 5548303. PMID 28629775.
- Fasolino M, Zhou Z (May 2017). "The Crucial Role of DNA Methylation and MeCP2 in Neuronal Function". Genes. 8 (5): 141. doi:10.3390/genes8050141. PMC 5448015. PMID 28505093.
- Bird A (January 2002). "DNA methylation patterns and epigenetic memory". Genes & Development. 16 (1): 6–21. doi:10.1101/gad.947102. PMID 11782440.
- Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017). "Experience-dependent epigenomic reorganization in the hippocampus". Learning & Memory. 24 (7): 278–288. doi:10.1101/lm.045112.117. PMC 5473107. PMID 28620075.
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, et al. (January 2016). "DNA methylation changes in plasticity genes accompany the formation and maintenance of memory". Nature Neuroscience. 19 (1): 102–10. doi:10.1038/nn.4194. PMC 4700510. PMID 26656643.
- Zhou X, Zhuang Z, Wang W, He L, Wu H, Cao Y, et al. (September 2016). "OGG1 is essential in oxidative stress induced DNA demethylation". Cellular Signalling. 28 (9): 1163–71. doi:10.1016/j.cellsig.2016.05.021. PMID 27251462.
- Day JJ, Sweatt JD (November 2010). "DNA methylation and memory formation". Nature Neuroscience. 13 (11): 1319–23. doi:10.1038/nn.2666. PMC 3130618. PMID 20975755.
- Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, et al. (May 2013). "Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β". Nature Neuroscience. 16 (5): 613–21. doi:10.1038/nn.3356. PMC 3637871. PMID 23525040.
- Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, et al. (June 2015). "Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes". Cell. 161 (7): 1592–605. doi:10.1016/j.cell.2015.05.032. PMC 4886855. PMID 26052046.
- Pérez-Cadahía B, Drobic B, Davie JR (February 2011). "Activation and function of immediate-early genes in the nervous system". Biochemistry and Cell Biology. 89 (1): 61–73. doi:10.1139/O10-138. PMID 21326363. S2CID 3257887.
- Colón-Cesario M, Wang J, Ramos X, García HG, Dávila JJ, Laguna J, et al. (May 2006). "An inhibitor of DNA recombination blocks memory consolidation, but not reconsolidation, in context fear conditioning". The Journal of Neuroscience. 26 (20): 5524–33. doi:10.1523/JNEUROSCI.3050-05.2006. PMC 6675301. PMID 16707804.
- Giglia-Mari G, Zotter A, Vermeulen W (January 2011). "DNA damage response". Cold Spring Harbor Perspectives in Biology. 3 (1): a000745. doi:10.1101/cshperspect.a000745. PMC 3003462. PMID 20980439.
- Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC (November 2012). "Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA". Nature. 491 (7423): 274–8. Bibcode:2012Natur.491..274B. doi:10.1038/nature11598. PMC 4112059. PMID 23103864.
- Erill I, Campoy S, Barbé J (2007). "Aeons of distress: an evolutionary perspective on the bacterial SOS response". FEMS Microbiol. Rev. 31 (6): 637–656. doi:10.1111/j.1574-6976.2007.00082.x. PMID 17883408.
- Murayama Y, Kurokawa Y, Mayanagi K, Iwasaki H (February 2008). "Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases". Nature. 451 (7181): 1018–21. Bibcode:2008Natur.451.1018M. doi:10.1038/nature06609. PMID 18256600. S2CID 205212254.
- Holthausen JT, Wyman C, Kanaar R (2010). "Regulation of DNA strand exchange in homologous recombination". DNA Repair (Amst). 9 (12): 1264–1272. doi:10.1016/j.dnarep.2010.09.014. PMID 20971042.
- Kathe SD, Shen GP, Wallace SS (April 2004). "Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts". The Journal of Biological Chemistry. 279 (18): 18511–20. doi:10.1074/jbc.M313598200. PMID 14978042.
- Brasnjevic I, Hof PR, Steinbusch HW, Schmitz C (2008). "Accumulation of nuclear DNA damage or neuron loss: molecular basis for a new approach to understanding selective neuronal vulnerability in neurodegenerative diseases". DNA Repair (Amst). 7 (7): 1087–1097. doi:10.1016/j.dnarep.2008.03.010. PMC 2919205. PMID 18458001.
- Hetman M, Vashishta A, Rempala G (2010). "Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition". J. Neurochem. 114 (6): 1537–1549. doi:10.1111/j.1471-4159.2010.06859.x. PMC 2945429. PMID 20557419.
- Piec I, Listrat A, Alliot J, Chambon C, Taylor RG, Bechet D (July 2005). "Differential proteome analysis of aging in rat skeletal muscle". FASEB Journal. 19 (9): 1143–5. doi:10.1096/fj.04-3084fje. PMID 15831715.
- Carnevale J, Palander O, Seifried LA, Dick FA (March 2012). "DNA damage signals through differentially modified E2F1 molecules to induce apoptosis". Molecular and Cellular Biology. 32 (5): 900–12. doi:10.1128/MCB.06286-11. PMC 3295199. PMID 22184068.
- Hittelman WN, Rao PN (1975). "Mutat. Res. 23 1974; 251; A.P. Rao and P.N. Rao, J. Natl. Cancer Inst. 57 1976; 1139; W.N. Hittelman and P.N. Rao, Cancer Res. 34 1974; 3433;". 35: 3027. Cite journal requires
|journal=(help) - Weinert TA, Hartwell LH (July 1988). "The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae". Science. 241 (4863): 317–22. Bibcode:1988Sci...241..317W. doi:10.1126/science.3291120. PMID 3291120. S2CID 36645009.
- Al-Moghrabi NM, Al-Sharif IS, Aboussekhra A (May 2001). "The Saccharomyces cerevisiae RAD9 cell cycle checkpoint gene is required for optimal repair of UV-induced pyrimidine dimers in both G(1) and G(2)/M phases of the cell cycle". Nucleic Acids Research. 29 (10): 2020–5. doi:10.1093/nar/29.10.2020. PMC 55462. PMID 11353070.