Matrix metalloproteinase
Matrix metalloproteinases (MMPs), also known as matrix metallopeptidases or matrixins, are metalloproteinases that are calcium-dependent zinc-containing endopeptidases;[1] other family members are adamalysins, serralysins, and astacins. The MMPs belong to a larger family of proteases known as the metzincin superfamily.[2]
| Matrix metalloproteinase | |
|---|---|
| Identifiers | |
| Symbol | MMP |
| Pfam clan | CL0126 |
| InterPro | IPR021190 |
| Membranome | 317 |
Collectively, these enzymes are capable of degrading all kinds of extracellular matrix proteins, but also can process a number of bioactive molecules. They are known to be involved in the cleavage of cell surface receptors, the release of apoptotic ligands (such as the FAS ligand), and chemokine/cytokine inactivation.[3] MMPs are also thought to play a major role in cell behaviors such as cell proliferation, migration (adhesion/dispersion), differentiation, angiogenesis, apoptosis, and host defense.
They were first described in vertebrates (1962),[4] including humans, but have since been found in invertebrates and plants. They are distinguished from other endopeptidases by their dependence on metal ions as cofactors, their ability to degrade extracellular matrix, and their specific evolutionary DNA sequence.
History
MMPs were described initially by Jerome Gross and Charles Lapiere (1962), who observed enzymatic activity (collagen triple helix degradation) during tadpole tail metamorphosis (by placing a tadpole tail in a collagen matrix plate).[5] Therefore, the enzyme was named interstitial collagenase (MMP-1).
Later, it was purified from human skin (1968),[6] and was recognized to be synthesized as a zymogen.[7]
The "cysteine switch" was described in 1990.[8]
Structure
The MMPs have a common domain structure. The three common domains are the pro-peptide, the catalytic domain, and the haemopexin-like C-terminal domain, which is linked to the catalytic domain by a flexible hinge region.[2]
The pro-peptide
The MMPs are initially synthesized as inactive zymogens with a pro-peptide domain that must be removed before the enzyme is active. The pro-peptide domain is part of the "cysteine switch." This contains a conserved cysteine residue that interacts with the zinc in the active site and prevents binding and cleavage of the substrate, keeping the enzyme in an inactive form. In the majority of the MMPs, the cysteine residue is in the conserved sequence PRCGxPD. Some MMPs have a prohormone convertase cleavage site (Furin-like) as part of this domain, which, when cleaved, activates the enzyme. MMP-23A and MMP-23B include a transmembrane segment in this domain.[9]
The catalytic domain
X-ray crystallographic structures of several MMP catalytic domains have shown that this domain is an oblate sphere measuring 35 x 30 x 30 Å (3.5 × 3 x 3 nm). The active site is a 20 Å (2 nm) groove that runs across the catalytic domain. In the part of the catalytic domain forming the active site there is a catalytically important Zn2+ ion, which is bound by three histidine residues found in the conserved sequence HExxHxxGxxH. Hence, this sequence is a zinc-binding motif.
The gelatinases, such as MMP-2, incorporate Fibronectin type II modules inserted immediately before in the zinc-binding motif in the catalytic domain.[10]
The hinge region
The catalytic domain is connected to the C-terminal domain by a flexible hinge or linker region. This is up to 75 amino acids long, and has no determinable structure.
The hemopexin-like C-terminal domain
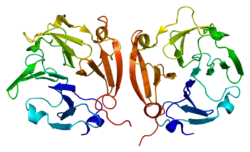
The C-terminal domain has structural similarities to the serum protein hemopexin. It has a four-bladed β-propeller structure. β-Propeller structures provide a large flat surface that is thought to be involved in protein–protein interactions. This determines substrate specificity and is the site for interaction with TIMP's (tissue inhibitor of metalloproteinases). The hemopexin-like domain is absent in MMP-7, MMP-23, MMP-26, and the plant and nematode. The membrane-bound MMPs (MT-MMPs) are anchored to the plasma membrane via a transmembrane or a GPI-anchoring domain.
Catalytic mechanism
There are three catalytic mechanisms published.
- In the first mechanism, Browner M.F. and colleagues[11] proposed the base-catalysis mechanism, carried out by the conserved glutamate residue and the Zn2+ ion.
- In the second mechanism, the Matthews-mechanism, Kester and Matthews[12] suggested an interaction between a water molecule and the Zn2+ ion during the acid-base catalysis.
- In the third mechanism, the Manzetti-mechanism, Manzetti Sergio and colleagues[13] provided evidence that a coordination between water and zinc during catalysis was unlikely, and suggested a third mechanism wherein a histidine from the HExxHxxGxxH-motif participates in catalysis by allowing the Zn2+ ion to assume a quasi-penta coordinated state, via its dissociation from it. In this state, the Zn2+ ion is coordinated with the two oxygen atoms from the catalytic glutamic acid, the substrate's carbonyl oxygen atom, and the two histidine residues, and can polarize the glutamic acid's oxygen atom, proximate the scissile bond, and induce it to act as reversible electron donor. This forms an oxyanion transition state. At this stage, a water molecule acts on the dissociated scissile bond and completes the hydrolyzation of the substrate.
Classification

The MMPs can be subdivided in different ways.
Evolutionary
Use of bioinformatic methods to compare the primary sequences of the MMPs suggest the following evolutionary groupings of the MMPs:
Analysis of the catalytic domains in isolation suggests that the catalytic domains evolved further once the major groups had differentiated, as is also indicated by the substrate specificities of the enzymes.
Functional
The most commonly used groupings (by researchers in MMP biology) are based partly on historical assessment of the substrate specificity of the MMP and partly on the cellular localization of the MMP. These groups are the collagenases, the gelatinases, the stromelysins, and the membrane-type MMPs (MT-MMPs).
- The collagenases are capable of degrading triple-helical fibrillar collagens into distinctive 3/4 and 1/4 fragments. These collagens are the major components of bone, cartilage and dentin, and MMPs are the only known mammalian enzymes capable of degrading them. The collagenases are No. 1, No. 8, No. 13, and No. 18. In addition, No. 14 has also been shown to cleave fibrillar collagen, and there is evidence that No. 2 is capable of collagenolysis. In MeSH, the current list of collagenases includes No. 1, No. 2, No. 8, No. 9, and No. 13. Collagenase No. 14 is present in MeSH but not listed as a collagenase, while No. 18 is absent from MeSH.
- The main substrates of the gelatinases are type IV collagen and gelatin, and these enzymes are distinguished by the presence of an additional domain inserted into the catalytic domain. This gelatin-binding region is positioned immediately before the zinc-binding motif, and forms a separate folding unit that does not disrupt the structure of the catalytic domain. The gelatinases are No. 2 and No. 9.
- The stromelysins display a broad ability to cleave extracellular matrix proteins but are unable to cleave the triple-helical fibrillar collagens. The three canonical members of this group are No. 3, No. 10, and No. 11.
- All six membrane-type MMPs (No. 14, No. 15, No. 16, No. 17, No. 24, and No. 25) have a furin cleavage site in the pro-peptide, which is a feature also shared by No. 11.
However, it is becoming increasingly clear that these divisions are somewhat artificial as there are a number of MMPs that do not fit into any of the traditional groups.
Genes
| Gene | Name | Aliases | Location | Description |
| MMP1 | Interstitial collagenase | CLG, CLGN | secreted | Substrates include Col I, II, III, VII, VIII, X, gelatin |
| MMP2 | Gelatinase-A, 72 kDa gelatinase | secreted | Substrates include Gelatin, Col I, II, III, IV, Vii, X | |
| MMP3 | Stromelysin 1 | CHDS6, MMP-3, SL-1, STMY, STMY1, STR1 | secreted | Substrates include Col II, IV, IX, X, XI, gelatin |
| MMP7 | Matrilysin, PUMP 1 | MMP-7, MPSL1, PUMP-1 | secreted | membrane associated through binding to cholesterol sulfate in cell membranes, substrates include: fibronectin, laminin, Col IV, gelatin |
| MMP8 | Neutrophil collagenase | CLG1, HNC, MMP-8, PMNL-CL | secreted | Substrates include Col I, II, III, VII, VIII, X, aggrecan, gelatin |
| MMP9 | Gelatinase-B, 92 kDa gelatinase | CLG4B, GELB, MANDP2, MMP-9 | secreted | Substrates include Gelatin, Col IV, V |
| MMP10 | Stromelysin 2 | SL-2, STMY2 | secreted | Substrates include Col IV, laminin, fibronectin, elastin |
| MMP11 | Stromelysin 3 | SL-3, ST3, STMY3 | secreted | MMP-11 shows more similarity to the MT-MMPs, is convertase-activatable and is secreted therefore usually associated to convertase-activatable MMPs. Substrates include Col IV, fibronectin, laminin, aggrecan |
| MMP12 | Macrophage metalloelastase | HME, ME, MME, MMP-12 | secreted | Substrates include elastin, fibronectin, Col IV |
| MMP13 | Collagenase 3 | CLG3, MANDP1, MMP-13 | secreted | Substrates include Col I, II, III, IV, IX, X, XIV, gelatin |
| MMP14 | MT1-MMP | MMP-14, MMP-X1, MT-MMP, MT-MMP 1, MT1-MMP, MT1MMP, MTMMP1, WNCHRS | membrane-associated | type-I transmembrane MMP; substrates include gelatin, fibronectin, laminin |
| MMP15 | MT2-MMP | MT2-MMP, MTMMP2, SMCP-2, MMP-15, MT2MMP | membrane-associated | type-I transmembrane MMP; substrates include gelatin, fibronectin, laminin |
| MMP16 | MT3-MMP | C8orf57, MMP-X2, MT-MMP2, MT-MMP3, MT3-MMP | membrane-associated | type-I transmembrane MMP; substrates include gelatin, fibronectin, laminin |
| MMP17 | MT4-MMP | MT4-MMP, MMP-17, MT4MMP, MTMMP4 | membrane-associated | glycosyl phosphatidylinositol-attached; substrates include fibrinogen, fibrin |
| MMP18 | Collagenase 4, xcol4, xenopus collagenase | – | No known human orthologue | |
| MMP19 | RASI-1, occasionally referred to as stromelysin-4 | MMP18, RASI-1, CODA | – | |
| MMP20 | Enamelysin | AI2A2, MMP-20 | secreted | |
| MMP21 | X-MMP | MMP-21, HTX7 | secreted | |
| MMP23A | CA-MMP | membrane-associated | type-II transmembrane cysteine array | |
| MMP23B | – | MIFR, MIFR-1, MMP22, MMP23A | membrane-associated | type-II transmembrane cysteine array |
| MMP24 | MT5-MMP | MMP-24, MMP25, MT-MMP 5, MT-MMP5, MT5-MMP, MT5MMP, MTMMP5 | membrane-associated | type-I transmembrane MMP |
| MMP25 | MT6-MMP | MMP-25, MMP20, MMP20A, MMPL1, MT-MMP 6, MT-MMP6, MT6-MMP, MT6MMP, MTMMP6 | membrane-associated | glycosyl phosphatidylinositol-attached |
| MMP26 | Matrilysin-2, endometase | – | ||
| MMP27 | MMP-22, C-MMP | MMP-27 | – | |
| MMP28 | Epilysin | EPILYSIN, MM28, MMP-25, MMP-28, MMP25 | secreted | Discovered in 2001 and given its name due to have been discovered in human keratinocytes. Unlike other MMPs this enzyme is constitutivley expressed in many tissues (Highly expressed in testis and at lower levels in lung, heart, brain, colon, intestine, placenta, salivary glands, uterus, skin). A threonine replaces proline in its cysteine switch (PRCGVTD).[14] |
Matrix metalloproteinases combines with the metal binding protein, metallothionine; thus helping in metal binding mechanism.
Function
The MMPs play an important role in tissue remodeling associated with various physiological or pathological processes such as morphogenesis, angiogenesis, tissue repair, cirrhosis, arthritis, and metastasis. MMP-2 and MMP-9 are thought to be important in metastasis. MMP-1 is thought to be important in rheumatoid arthritis and osteoarthritis. Recent data suggests active role of MMPs in the pathogenesis of Aortic Aneurysm. Excess MMPs degrade the structural proteins of the aortic wall. Disregulation of the balance between MMPs and TIMPs is also a characteristic of acute and chronic cardiovascular diseases.[15]
Activation
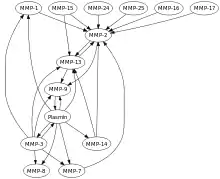
All MMPs are synthesized in the latent form (Zymogen). They are secreted as proenzymes and require extracellular activation. They can be activated in vitro by many mechanisms including organomercurials, chaotropic agents, and other proteases.
Inhibitors
The MMPs are inhibited by specific endogenous tissue inhibitor of metalloproteinases (TIMPs), which comprise a family of four protease inhibitors: TIMP-1, TIMP-2, TIMP-3, and TIMP-4.
Synthetic inhibitors generally contain a chelating group that binds the catalytic zinc atom at the MMP active site tightly. Common chelating groups include hydroxamates, carboxylates, thiols, and phosphinyls. Hydroxymates are particularly potent inhibitors of MMPs and other zinc-dependent enzymes, due to their bidentate chelation of the zinc atom. Other substituents of these inhibitors are usually designed to interact with various binding pockets on the MMP of interest, making the inhibitor more or less specific for given MMPs.[2]
Pharmacology
Doxycycline, at subantimicrobial doses, inhibits MMP activity, and has been used in various experimental systems for this purpose, such as for recalcitrant recurrent corneal erosions. It is used clinically for the treatment of periodontal disease and is the only MMP inhibitor that is widely available clinically. It is sold under the trade name Periostat by the company CollaGenex. Minocycline, another tetracycline antibiotic, has also been shown to inhibit MMP activity.
A number of rationally designed MMP inhibitors have shown some promise in the treatment of pathologies that MMPs are suspected to be involved in (see above). However, most of these, such as marimastat (BB-2516), a broad-spectrum MMP inhibitor, and cipemastat (Ro 32-3555), an MMP-1 selective inhibitor, have performed poorly in clinical trials. The failure of Marimastat was partially responsible for the folding of British Biotech, which developed it. The failure of these drugs has been due largely to toxicity (in particular, musculo-skeletal toxicity in the case of broad spectrum inhibitors) and failure to show expected results (in the case of trocade, promising results in rabbit arthritis models were not replicated in human trials). The reasons behind the largely disappointing clinical results of MMP inhibitors is unclear, especially in light of their activity in animal models.
See also
- Proteases in angiogenesis
- Drug discovery and development of MMP inhibitors
- Collagen Hybridizing Peptide, a peptide that can bind and stain MMP cleaved collagen
References
- Verma RP, Hansch C (March 2007). "Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs" (PDF). Bioorg. Med. Chem. 15 (6): 2223–68. doi:10.1016/j.bmc.2007.01.011. PMID 17275314. Archived from the original (PDF) on 13 May 2015. Retrieved 21 October 2015.
- Matrix Metalloproteinases: Its implications in cardiovascular disorders
- Van Lint P, Libert C (December 2007). "Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation". J. Leukoc. Biol. 82 (6): 1375–81. doi:10.1189/jlb.0607338. PMID 17709402.
- Gross, J.; Lapiere, C. M. (June 1962). "Collagenolytic activity in amphibian tissues: a tissue culture assay". Proceedings of the National Academy of Sciences. 48 (6): 1014–22. Bibcode:1962PNAS...48.1014G. doi:10.1073/pnas.48.6.1014. PMC 220898. PMID 13902219.
- Gross J, Lapiere C (1962). "Collagenolytic Activity in Amphibian Tissues: A Tissue Culture Assay". Proc Natl Acad Sci USA. 48 (6): 1014–22. Bibcode:1962PNAS...48.1014G. doi:10.1073/pnas.48.6.1014. PMC 220898. PMID 13902219.
- Eisen A, Jeffrey J, Gross J (1968). "Human skin collagenase. Isolation and mechanism of attack on the collagen molecule". Biochim Biophys Acta. 151 (3): 637–45. doi:10.1016/0005-2744(68)90010-7. PMID 4967132.
- Harper E, Bloch K, Gross J (1971). "The zymogen of tadpole collagenase". Biochemistry. 10 (16): 3035–41. doi:10.1021/bi00792a008. PMID 4331330.
- Van Wart H, Birkedal-Hansen H (1990). "The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family". Proc Natl Acad Sci USA. 87 (14): 5578–82. Bibcode:1990PNAS...87.5578V. doi:10.1073/pnas.87.14.5578. PMC 54368. PMID 2164689.
- Pei D, Kang T, Qi H (2000). "Cysteine array matrix metalloproteinase (CA-MMP)/MMP-23 is a type II transmembrane matrix metalloproteinase regulated by a single cleavage for both secretion and activation". J Biol Chem. 275 (43): 33988–97. doi:10.1074/jbc.M006493200. PMID 10945999.
- Trexler M, Briknarová K, Gehrmann M, Llinás M, Patthy L (2003). "Peptide ligands for the fibronectin type II modules of matrix metalloproteinase 2 (MMP-2)". J Biol Chem. 278 (14): 12241–6. doi:10.1074/jbc.M210116200. PMID 12486137.
- Browner MF, Smith WW, Castelhano AL (1995). "Matrilysin-inhibitor complexes: common themes among metalloproteases". Biochemistry. 34 (20): 6602–10. doi:10.1021/bi00020a004. PMID 7756291.
- Kester WR, Matthews BW (1977). "Crystallographic study of the binding of dipeptide inhibitors to thermolysin: implications for the mechanism of catalysis". Biochemistry. 16 (11): 2506–16. doi:10.1021/bi00630a030. PMID 861218.
- Manzetti S, McCulloch DR, Herington AC, van der Spoel D (2003). "Modeling of enzyme-substrate complexes for the metalloproteases MMP-3, ADAM-9 and ADAM-10". J. Comput.-Aided Mol. Des. 17 (9): 551–65. Bibcode:2003JCAMD..17..551M. doi:10.1023/B:JCAM.0000005765.13637.38. PMID 14713188. S2CID 17453639.
- Lohi J, Wilson CL, Roby JD, Parks WC (2001). "Epilysin, a novel human matrix metalloproteinase (MMP-28) expressed in testis and keratinocytes and in response to injury". J Biol Chem. 276 (13): 10134–10144. doi:10.1074/jbc.M001599200. PMID 11121398.
- Snoek-van Beurden PAM; Von den Hoff JW (2005). "Zymographic techniques for the analysis of matrix metalloproteinases and their inhibitors". BioTechniques. 38 (1): 73–83. doi:10.2144/05381RV01. PMID 15679089.
Synergistic effect of stromelysin-1 (matrix metalloproteinase-3) promoter (-1171 5A->6A) polymorphism in oral submucous fibrosis and head and neck lesions.Chaudhary AK, Singh M, Bharti AC, Singh M, Shukla S, Singh AK, Mehrotra R. BMC Cancer. 2010 Jul 14;10:369.
External links
- MBInfo – Matrix metalloproteinases (MMPs) facilitate extracellular matrix disassembly
- The Matrix Metalloproteinase Protein
- Extracellular proteolysis at fibrinolysis.org
- Currently identified substrates for mammalian MMPs at clip.ubc.ca
- Matrix+metalloproteinases at the US National Library of Medicine Medical Subject Headings (MeSH)