Terrabacteria
Terrabacteria is a taxon containing approximately two-thirds (6,157 sp.) of prokaryote species, including those in the gram positive phyla (Actinobacteria and Firmicutes) as well as the phyla Cyanobacteria, Chloroflexi, and Deinococcus-Thermus.[1][2]
| Terrabacteria | |
|---|---|
 | |
| Scanning electron micrograph of Actinomyces israelii (Actinobacteria) | |
| Scientific classification | |
| Domain: | |
| (unranked): | Terrabacteria Battistuzzi et al., 2004, Battistuzzi & Hedges, 2009 |
| Phyla | |
| |
| Synonyms | |
| |
It derives its name (terra = "land") from the evolutionary pressures of life on land. Terrabacteria possess important adaptations such as resistance to environmental hazards (e.g., desiccation, ultraviolet radiation, and high salinity) and oxygenic photosynthesis. Also, the unique properties of the cell wall in gram-positive taxa, which likely evolved in response to terrestrial conditions, have contributed toward pathogenicity in many species.[2] These results now leave open the possibility that terrestrial adaptations may have played a larger role in prokaryote evolution than currently understood.[1][2]
Terrabacteria was proposed in 2004 for Actinobacteria, Cyanobacteria, and Deinococccus-Thermus [1] and was expanded later to include Firmicutes and Chloroflexi.[2] Other phylogenetic analyses [3] have supported the close relationships of these phyla. Most species of prokaryotes not placed in Terrabacteria were assigned to the taxon Hydrobacteria [2] (3,203 sp.), in reference to the moist environment inferred for the common ancestor of those species. Terrabacteria and Hydrobacteria were inferred to have diverged approximately 3 billion years ago, suggesting that land (continents) had been colonized by prokaryotes at that time.[2] Together, Terrabacteria and Hydrobacteria form a large group containing 99% (9,360 sp.) of all Eubacteria known by 2009, and placed in the taxon Selabacteria, in allusion to their phototrophic abilities (selas = light).[4]
Terrabacteria should not be confused with the recently described taxon "Glidobacteria",[5] which includes only some members of Terrabacteria but excludes Firmicutes and Actinobacteria, and is not supported by molecular phylogenetic data.[2]
Phylogeny
The phylogenetic tree according to the phylogenetic analyzes of Battistuzzi (2009) is the following and with a molecular clock calibration.[1][2]
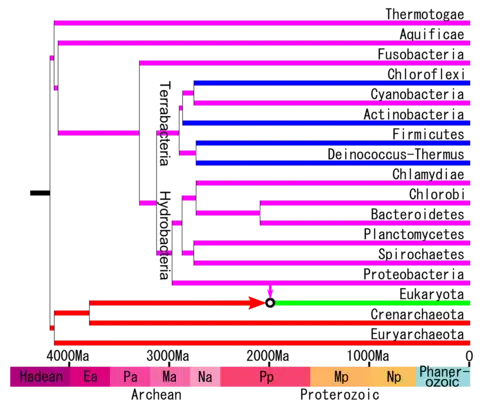
Recent molecular analyzes have found roughly the following relationships including other phyla, whose relationships were uncertain.[6][7][8][9][10][11]
| Terrabacteria |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
On the other hand, Coleman et al (2020) named the clade composed of Thermotogae, Deinococcus-Thermus, Synergistetes and related as DST and furthermore the analysis suggests that ultra-small bacteria (CPR group) may belong to Terrabacteria being more closely related to Chloroflexi. According to this study the phylum Aquificae sometimes included belongs to Gracilicutes and that the phylum Fusobacteria can belong to both Terrabacteria and Gracilicutes. The result was the following:[12]
| Terrabacteria |
| ||||||||||||||||||||||||||||||||||||||||||||||||
It is possible that Terrabacteria is also a paraphyletic clade according to other phylogenetic analyzes.[13][14]
References
- Battistuzzi, F. U.; Feijão, A.; Hedges, S. B. (2004). "A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land". BMC Evolutionary Biology. 4: 44. doi:10.1186/1471-2148-4-44. PMC 533871. PMID 15535883.
- Battistuzzi, FU; Hedges, SB (2009). "A major clade of prokaryotes with ancient adaptations to life on land". Molecular Biology and Evolution. 26 (2): 335–43. doi:10.1093/molbev/msn247. PMID 18988685.
- Bern, M; Goldberg, D (2005). "Automatic selection of representative proteins for bacterial phylogeny". BMC Evolutionary Biology. 5 (1): 34. doi:10.1186/1471-2148-5-34. PMC 1175084. PMID 15927057.
- Battistuzzi, F. U., Hedges, S. B. 2009. Eubacteria. Pp. 106-115 in The Timetree of Life, S. B. Hedges and S. Kumar, Eds. (Oxford University Press, New York, 2009). http://www.timetree.org/book.php.
- Cavalier-Smith, T (2006). "Rooting the tree of life by transition analyses". Biology Direct. 1 (1): 19. doi:10.1186/1745-6150-1-19. PMC 1586193. PMID 16834776.
- Karthik Anantharaman et al. 2016, Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system Nature Communications DOI: 10.1038/ncomms13219
- Paula B. Matheus Carnevali et al. 2019, Hydrogen-based metabolism as an ancestral trait inlineages sibling to the Cyanobacteria. Nature Communications 10(1) · December 2019 DOI: 10.1038/s41467-018-08246-y
- Ji, M., Greening, C., Vanwonterghem, I. et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552, 400–403 (2017) doi:10.1038/nature25014
- Tahon G. et al. 2018, Abditibacterium utsteinense sp. nov., the first cultivated member of candidate phylum FBP, isolated from ice-free Antarctic soil samples. Syst Appl Microbiol. 2018 Jul;41(4):279-290. doi: 10.1016/j.syapm.2018.01.009. Epub 2018 Feb 7.
- Christian Rinke et al 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature Volume: 499, Pages: 431–437 doi:10.1038/nature12352
- Emiley A Eloe-Fadrosh et al. 2017, Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs. Nature Communications 7 · January 2016 DOI: 10.1038/ncomms1047
- Gareth A. Coleman, Adrián A. Davín, Tara Mahendrarajah, Anja Spang, Philip Hugenholtz, Gergely J. Szöllősi, Tom A. Williams. (2020). A rooted phylogeny resolves early bacterial evolution. Biorxiv.
- Hug, L. A. et al. 2016, A new view of the tree of life. Nature Microbiology, 1, 16048.
- Zhu, Q., Mai, U., Pfeiffer, W. et al. (2019) «Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea». Nature Communications, 10: 5477