Epsilonproteobacteria
Epsilonproteobacteria are a class of Proteobacteria.[1] All species of this class are, like all Proteobacteria, Gram-negative.
| Epsilonproteobacteria | |
|---|---|
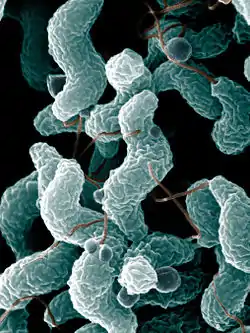 | |
| Campylobacter | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Proteobacteria |
| Class: | Epsilonproteobacteria |
| Orders | |
| |
The Epsilonproteobacteria consist of few known genera, mainly the curved to spirilloid Wolinella spp., Helicobacter spp., and Campylobacter spp. Most of the known species inhabit the digestive tracts of animals and serve as symbionts (Wolinella spp. in cattle) or pathogens (Helicobacter spp. in the stomach, Campylobacter spp. in the duodenum). Many Epsilonproteobacteria are motile with flagella.[2]
Numerous environmental sequences and isolates of Epsilonproteobacteria have also been recovered from hydrothermal vents and cold seep habitats. Examples of isolates include Sulfurimonas autotrophica,[3] Sulfurimonas paralvinellae,[4] Sulfurovum lithotrophicum[5] and Nautilia profundicola.[6] A member of the class Epsilonproteobacteria occurs as an endosymbiont in the large gills of the deepwater sea snail Alviniconcha hessleri.[7]
The Epsilonproteobacteria found at deep-sea hydrothermal vents characteristically exhibit chemolithotrophy, meeting their energy needs by oxidizing reduced sulfur, formate, or hydrogen coupled to the reduction of nitrate or oxygen.[8] Autotrophic Epsilonproteobacteria use the reverse Krebs cycle to fix carbon dioxide into biomass, a pathway originally thought to be of little environmental significance. The oxygen sensitivity of this pathway is consistent with their microaerophilic or anaerobic niche in these environments, and their likely evolution in the Mesoproterozoic oceans,[9] which are thought to have been sulfidic with low levels of oxygen available from cyanobacterial photosynthesis.[10]
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) [11] and National Center for Biotechnology Information (NCBI)[12] and the phylogeny is based on 16S rRNA-based LTP release 106 by 'The All-Species Living Tree' Project [13]
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Notes:
- Prokaryotes where no pure (axenic) cultures are isolated or available, i.e. not cultivated or can not be sustained in culture for more than a few serial passages
References
- "www.ncbi.nlm.nih.gov". Retrieved 2009-03-19.
- Beeby, M (December 2015). "Motility in the epsilon-proteobacteria". Current Opinion in Microbiology. 28: 115–21. doi:10.1016/j.mib.2015.09.005. hdl:10044/1/27763. PMID 26590774.
- Inagaki, F. (2003-11-01). "Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing -proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough". International Journal of Systematic and Evolutionary Microbiology. 53 (6): 1801–1805. doi:10.1099/ijs.0.02682-0. ISSN 1466-5026. PMID 14657107.
- Takai, K. (2006-08-01). "Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas". International Journal of Systematic and Evolutionary Microbiology. 56 (8): 1725–1733. doi:10.1099/ijs.0.64255-0. ISSN 1466-5026. PMID 16901999.
- Inagaki, Fumio; Ken Takai; Kenneth H. Nealson; Koki Horikoshi (2004-09-01). "Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments". International Journal of Systematic and Evolutionary Microbiology. 54 (5): 1477–1482. doi:10.1099/ijs.0.03042-0. ISSN 1466-5026. PMID 15388698.
- Julie L. Smith; Barbara J. Campbell; Thomas E. Hanson; Chuanlun L. Zhang; S. Craig Cary (2008). "Nautilia profundicola sp. nov., a thermophilic, sulfur-reducing epsilonproteobacterium from deep-sea hydrothermal vents". International Journal of Systematic and Evolutionary Microbiology. 58 (7): 1598–1602. doi:10.1099/ijs.0.65435-0. PMID 18599701.
- Suzuki, Yohey; Sasaki, Takenori; Suzuki, Masae; Nogi, Yuichi; Miwa, Tetsuya; Takai, Ken; Nealson, Kenneth H.; Horikoshi, Koki (2005). "Novel Chemoautotrophic Endosymbiosis between a Member of the Epsilonproteobacteria and the Hydrothermal-Vent Gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean". Applied and Environmental Microbiology. 71 (9): 5440–5450. doi:10.1128/AEM.71.9.5440-5450.2005. PMC 1214688. PMID 16151136.
- Takai, Ken; et al. (2005). "Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria" (PDF). Applied and Environmental Microbiology. 71 (11): 7310–7320. doi:10.1128/aem.71.11.7310-7320.2005. PMC 1287660. PMID 16269773.
- Campbell, Barbara J.; Annette Summers Engel; Megan L. Porter; Ken Takai (2006-05-02). "The versatile ε-proteobacteria: key players in sulphidic habitats". Nature Reviews Microbiology. 4 (6): 458–468. doi:10.1038/nrmicro1414. ISSN 1740-1526. PMID 16652138. S2CID 10479314.
- Anbar, A. D.; A. H. Knoll (2002-08-16). "Proterozoic Ocean Chemistry and Evolution: A Bioinorganic Bridge?". Science. 297 (5584): 1137–1142. Bibcode:2002Sci...297.1137A. CiteSeerX 10.1.1.615.3041. doi:10.1126/science.1069651. PMID 12183619. S2CID 5578019.
- J.P. Euzéby. "Epsilonproteobacteria". List of Prokaryotic names with Standing in Nomenclature (LPSN). Archived from the original on 2011-10-07. Retrieved 2011-11-17.
- Sayers; et al. "Epsilonproteobacteria". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2011-06-05.
- 'The All-Species Living Tree' Project."16S rRNA-based LTP release 106 (full tree)" (PDF). Silva Comprehensive Ribosomal RNA Database. Retrieved 2011-11-17.
External links
- Epsilonproteobacteria at the US National Library of Medicine Medical Subject Headings (MeSH)