Teseptimavirus
Teseptimavirus (synonyms T7 phage group, T7-like phages, T7-like viruses, T7likevirus) is a genus of viruses in the order Caudovirales, in the family Autographiviridae, in the subfamily Studiervirinae. Bacteria serve as the natural host, with transmission achieved through passive diffusion. There are currently 17 species in this genus, including the type species Escherichia virus T7.[1][2][3]
| Teseptimavirus | |
|---|---|
 | |
| Escherichia virus T7 | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Uroviricota |
| Class: | Caudoviricetes |
| Order: | Caudovirales |
| Family: | Autographiviridae |
| Subfamily: | Studiervirinae |
| Genus: | Teseptimavirus |
| Type species | |
| Escherichia virus T7 | |
Taxonomy
The following species are recognized:[2]
- Enterobacteria virus IME390
- Escherichia virus 13a
- Escherichia virus 64795ec1
- Escherichia virus C5
- Escherichia virus CICC80001
- Escherichia virus Ebrios
- Escherichia virus EG1
- Escherichia virus HZ2R8
- Escherichia virus HZP2
- Escherichia virus N30
- Escherichia virus NCA
- Escherichia virus T7
- Salmonella virus 3A8767
- Salmonella virus Vi06
- Stenotrophomonas virus IME15
- Yersinia virus YpPY
- Yersinia virus YpsPG
Structure
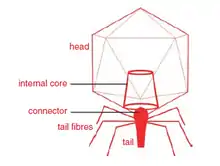
Sp6likeviruses are nonenveloped, with a head and tail. The head has icosahedral symmetry (T=7) approximately 60 nm in diameter. The tail is non-contractile and has six short subterminal fibers.[1]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Teseptimavirus | Head-Tail | T=7 | Non-enveloped | Linear | Monopartite |
Genome
Genomes are linear, around 40-42kb in length.[1] All three viruses' genomes have been fully sequenced and are available on NCBI's website. They range between 37k and 40k nucleotides, with 42 to 60 proteins. All three complete genomes, as well as several additional "unclassified" virus genomes, are available here[3]
Life cycle
Viral replication is cytoplasmic. The virus attaches to the host cell adhesion receptors using its tail fibers, and ejects the viral DNA into the host periplasm via short tail system. Class I genes are transcribed by the host cell's RNA polymerase before the viral genome completely exits the capsid. Class II genes are then transcribed by T7 RNA polymerase and the host genome is degraded. Genomic DNA is replicated by T7 DNA polymerase and concatemers are formed before Class III genes are transcribed. Finally, the procapsid is assembled and packed, the tail is assembled and the mature virions are released via lysis. Bacteria serve as the natural host. Transmission routes are passive diffusion.[1]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Teseptimavirus | Bacteria | None | Injection | Lysis | Cytoplasm | Cytoplasm | Passive diffusion |
History
According to ICTV's second report in 1976, the genus T7likevirus was first accepted under the name T7 phage group, unassigned to an order, family, or sub-family. In ICTV's third report in 1981, the genus was classified into the family Poloviridae. T7 phage group was renamed to T7-like phages in the sixth report in 1995. In 1998, the whole family was moved into the newly created order Caudovirales, and the genus was renamed again in the seventh report in 1999 to T7-like viruses. 2009 saw the genus moved into the newly created subfamily Autographivirinae, and it was renamed again in 2012 to T7likevirus.[2] The genus was later renamed to Teseptimavirus under the newly established family Autographiviridae and subfamily Studiervirinae.
References
- "Viral Zone". ExPASy. Retrieved 17 February 2015.
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 4 May 2020.
- NCBI. "T7linkevirus Complete Genomes". Retrieved 17 February 2015.