Caulimoviridae
Caulimoviridae is a family of viruses infecting plants.[1] There are currently 85 species in this family, divided among 10 genera.[2] Viruses belonging to the family Caulimoviridae are termed double-stranded DNA (dsDNA) reverse-transcribing viruses (or pararetroviruses) i.e. viruses that contain a reverse transcription stage in their replication cycle. This family contains all plant viruses with a dsDNA genome that have a reverse transcribing phase in its lifecycle.
| Caulimoviridae | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Ortervirales |
| Family: | Caulimoviridae |
Taxonomy
The following genera are recognized:
- Badnavirus
- Caulimovirus
- Cavemovirus
- Dioscovirus
- Petuvirus
- Rosadnavirus
- Solendovirus
- Soymovirus
- Tungrovirus
- Vaccinivirus
Virus particle structure
All viruses of this family are non-enveloped. Virus particles are either bacilliform or isometric. The type of nucleocapsid incorporated into the virus structure determines the size of the viral particles. Bacilliform particles are approximately 35–50 nm in diameter and up to 900 nm in length. Isometric particles are on average 45–50 nm in diameter and show icosahedral symmetry.
Genome structure and replication
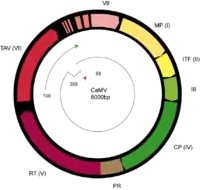
The genomes of viruses from this family contain monopartite, non-covalently closed circular dsDNA of 7.2–9.3 kbp with discontinuities in both genome strands at specific places. These genomes contain one open reading frame (ORF), as observed in petuviruses, to eight ORFs such as in the soymoviruses. Proteins encoded by the viral genomes include reverse transcriptase-ribonuclease H, aspartic proteases, nucleocapsids and transactivators — there are other proteins essential for replication that have yet to be assigned a specific function.
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Rosadnavirus | Icosahedral | T=7 | Non-enveloped | Circular | Monopartite |
| Cavemovirus | Icosahedral | T=7 | Non-enveloped | Circular | Monopartite |
| Petuvirus | Icosahedral | T=7 | Non-enveloped | Circular | Monopartite |
| Caulimovirus | Icosahedral | T=7 | Non-enveloped | Circular | Monopartite |
| Soymovirus | Icosahedral | T=7 | Non-enveloped | Circular | Monopartite |
| Badnavirus | Bacilliform | T=3 | Non-enveloped | Circular | Monopartite |
| Solendovirus | Icosahedral | T=7 | Non-enveloped | Circular | Monopartite |
| Tungrovirus | Bacilliform | T=3 | Non-enveloped | Circular | Monopartite |
Replication takes place in both the cytoplasm and the nucleus of host cells. Firstly, the viral genome enters the cytoplasm. The viral DNA forms supercoiled mini-chromosome structures upon entering the host nucleus, where it is transcribed into polyadenylated RNA which is terminally redundant (due to transcription occurring twice for some parts of the DNA). Newly transcribed RNA enters the cytoplasm where it is either translated into viral proteins, or retrotranscribed into new copies of the dsDNA viral genome by the viral reverse transcriptase. New dsDNA genomes are encapsidated in the cytoplasm and released.
The replication process involves a retro transcription step and an RNA intermediate, therefore viruses from the family Caulimoviridae are not considered true dsDNA viruses — instead they are termed DNA reverse-transcribing viruses. They share this characteristic with retroviruses. However, unlike retroviruses, viruses from the family Caulimoviridae do not require the integration of the viral genome into the genome of their hosts in order to replicate and for this reason their genome does not encode the enzymatic protein integrase.
The presence of endogenous viral elements (EVEs) in plant genomes is widespread.[3][4][5] and most known plant EVEs originate from viruses with DNA genomes in the family Caulimoviridae. Integration is thought to occur through non-homologous end-joining (illegitimate recombination) during DNA repair mechanisms. Most plant EVEs are non infectious. However, infectious Caulimoviridae EVEs have been reported in the genome of petunia [6](Petunia vein clearing virus), banana [7](Banana streak OL virus, Banana streak GF virus, Banana streak IM virus) and Nicotiana edwardsonii [8](Tobacco vein clearing virus).
| Genus | Host details | Tissue tropism | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|
| Rosadnavirus | Plants | None | Nucleus | Cytoplasm | Aphids |
| Cavemovirus | Plants | None | Nucleus | Cytoplasm | Aphids |
| Petuvirus | Plants | None | Nucleus | Cytoplasm | Grafting, activation of infectious EVEs |
| Caulimovirus | Plants | None | Nucleus | Cytoplasm | Aphids |
| Soymovirus | Plants | None | Nucleus | Cytoplasm | Aphids |
| Badnavirus | Plants | None | Nucleus | Cytoplasm | Mealybugs, activation of infectious EVEs |
| Solendovirus | Plants | None | Nucleus | Cytoplasm | Activation of infectious EVEs |
| Tungrovirus | Plants | None | Nucleus | Cytoplasm | Aphids |
References
- Teycheney, PY; Geering, ADW; Dasgupta, I; Hull, R; Kreuze, JF; Lockhart, B; Muller, E; Olszewski, N; Pappu, H; Pooggin, MM; Richert-Pöggeler, KR; Schoelz, JE; Seal, S; Stavolone, L; Umber, M; ICTV Report Consortium (October 2020). "ICTV Virus Taxonomy Profile: Caulimoviridae". The Journal of General Virology. 101 (10): 1025–1026. doi:10.1099/jgv.0.001497. PMID 32940596.
- "ICTV Report Caulimoviridae".
- Recent advances in plant virology. Caranta, Carole. Norfolk, UK: Caister Academic Press. 2011. ISBN 9781904455752. OCLC 644654169.CS1 maint: others (link)
- Geering, Andrew D. W.; Maumus, Florian; Copetti, Dario; Choisne, Nathalie; Zwickl, Derrick J.; Zytnicki, Matthias; McTaggart, Alistair R.; Scalabrin, Simone; Vezzulli, Silvia (10 November 2014). "Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution". Nature Communications. 5 (1): 5269. Bibcode:2014NatCo...5.5269G. doi:10.1038/ncomms6269. ISSN 2041-1723. PMC 4241990. PMID 25381880.
- Diop, Seydina Issa; Geering, Andrew D. W.; Alfama-Depauw, Françoise; Loaec, Mikaël; Teycheney, Pierre-Yves; Maumus, Florian (12 January 2018). "Tracheophyte genomes keep track of the deep evolution of the Caulimoviridae". Scientific Reports. 8 (1): 572. Bibcode:2018NatSR...8..572D. doi:10.1038/s41598-017-16399-x. ISSN 2045-2322. PMC 5766536. PMID 29330451.
- Richert‐Pöggeler, Katja R.; Noreen, Faiza; Schwarzacher, Trude; Harper, Glyn; Hohn, Thomas (15 September 2003). "Induction of infectious petunia vein clearing (pararetro) virus from endogenous provirus in petunia". The EMBO Journal. 22 (18): 4836–4845. doi:10.1093/emboj/cdg443. ISSN 0261-4189. PMC 212712. PMID 12970195.
- Gayral, Philippe; Noa-Carrazana, Juan-Carlos; Lescot, Magali; Lheureux, Fabrice; Lockhart, Benham E. L.; Matsumoto, Takashi; Piffanelli, Pietro; Iskra-Caruana, Marie-Line (1 July 2008). "A Single Banana Streak Virus Integration Event in the Banana Genome as the Origin of Infectious Endogenous Pararetrovirus". Journal of Virology. 82 (13): 6697–6710. doi:10.1128/JVI.00212-08. ISSN 0022-538X. PMC 2447048. PMID 18417582.
- Lockhart, B. E.; Menke, J.; Dahal, G.; Olszewski, N. E. (2000). "Characterization and genomic analysis of tobacco vein clearing virus, a plant pararetrovirus that is transmitted vertically and related to sequences integrated in the host genome". Journal of General Virology. 81 (6): 1579–1585. doi:10.1099/0022-1317-81-6-1579. PMID 10811941.
External links
- ICTV Report: Caulimoviridae
- "Caulimoviridae". Viralzone.
- Spumavirus at the US National Library of Medicine Medical Subject Headings (MeSH)
- "Caulimoviridae". NCBI Taxonomy Browser. 186534.