Heme oxygenase
Heme oxygenase or haem oxygenase (HO) is an enzyme that catalyzes the degradation of heme. This produces biliverdin, ferrous iron, and carbon monoxide.[1][2] HO was first described in the late 1960s when Raimo Tenhunen demonstrated an enzymatic reaction for heme catabolism.[3] HO is the premier source for endogenous carbon monoxide (CO) production. Indeed, monitored small doses of CO are being studied for therapeutic benefits.[4]
| heme oxygenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.14.99.3 | ||||||||
| CAS number | 9059-22-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Heme oxygenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structures of ferrous and ferrous-no forms of verdoheme in a complex with human heme oxygenase-1: catalytic implications for heme cleavage | |||||||||
| Identifiers | |||||||||
| Symbol | Heme_oxygenase | ||||||||
| Pfam | PF01126 | ||||||||
| Pfam clan | CL0230 | ||||||||
| InterPro | IPR016053 | ||||||||
| PROSITE | PDOC00512 | ||||||||
| SCOP2 | 1qq8 / SCOPe / SUPFAM | ||||||||
| Membranome | 532 | ||||||||
| |||||||||
Heme oxygenase
Heme oxygenase is a heme-containing member of the heat shock protein (HSP) family identified as HSP32. HO-1 is a 32kDa enzyme which contains 288 amino acid residues.[5] HO is located in the endoplasmic reticulum, though it has also been reported in the mitochondria, cell nucleus, and plasma membrane.[6]
HO catalyzes the degradation of heme to biliverdin/bilirubin, ferrous iron, and carbon monoxide. Though present throughout the body, HO has significant activity in the spleen in the degradation of hemoglobin during erythrocyte recycling (0.8% of the erythrocyte pool per day), which accounts for ~80% of heme derived endogenous CO production. The remaining 20% of heme derived CO production is largely attributed to hepatic catabolism of hemoproteins (myoglobin, cytochromes, catalase, peroxidases, soluble guanylate cyclase, nitric oxide synthase) and ineffective erythropoiesis in bone marrow.[7] HO enzymes are degraded via ubiquitination.[8] In humans three isoforms of heme oxygenase are known.
Heme oxygenase 1
Heme oxygenase 1 (HO-1) is a stress-induced isoform present throughout the body with highest concentrations in the spleen, liver, and kidneys.[9] HO-1 is a 32kDa enzyme containing 288 amino acid residues which is encoded by the HMOX1 gene. A study has found levels of HO-1 in lung tissue were directly related to infection with tuberculosis or infection-free areas, and knockout mice were found susceptible, showing the essential role of this enzyme.[10] HO-1 protects cells by reducing superoxide and other reactive oxygen species.[11]
Heme oxygenase 2
Heme oxygenase 2 (HO-2) is a constitutive isoform that is expressed under homeostatic conditions in the testes, endothelial cells and the brain.[12] HO-2 is encoded by the HMOX2 gene. HO-2 is 36 kDa and shares 47% similarity with the HO-1 amino acid sequence.
Heme oxygenase 3
A third heme oxygenase (HO-3) is considered to be catalytically inactive and is thought to work in heme sensing or heme binding. HO-3 is 33 kDa with greatest presence in the liver, prostate, and kidneys.[9]
Microbial heme oxygenase
Heme oxygenase is conserved across phylogenetic kingdoms.[13] The European Bioinformatics Institute's InterPro taxonomy database indicates there are 4,347 bacteria species, 552 fungi species, and 6 archaea species expressing a HO-1-like enzymes. Microbial HO homologues use different abbreviation such as HMX1 in Saccharomyces cerevisiae,[14] Hmu O in Corynebacterium diphtheriae,[15] and Chu S in Escherichia coli.[16] A critical role of the prokaryotic HO systems is to facilitate acquisition of nutritional iron from a eukaryotic host.[17] Some HO-like prokaryotic enzymes are inactive or do not liberate CO. Certain strains of Escherichia coli express the non-CO producing Chu W isoform, whilst HO-like enzymes in other microbes have been reported to produce formaldehyde.[18][19]
The human microbiome contributes to endogenous carbon monoxide production in humans.[20]
Reaction
Heme oxygenase cleaves the heme ring at the alpha-methene bridge to form either biliverdin or, if the heme is still attached to a globin, verdoglobin. Biliverdin is subsequently converted to bilirubin by biliverdin reductase. The reaction comprises three steps, which may be:[21]
- Heme b3+ + O
2 + NADPH + H+
→ α-meso-hydroxyheme3+ + NADP+
+ H
2O - α-meso-hydroxyheme3+ + H+
+ O
2 → verdoheme4+ + CO + H2O - verdoheme4+ + 7/2 NADPH + O
2+ 3/2 H+
→ biliverdin + Fe2+ + 7/2 NADP+
+ H
2O
- Heme b3+ + O
The sum of these reactions is:
- Heme b3+ + 3O
2 + 9/2 NADPH + 7/2 H+
→ biliverdin + Fe2+ + CO + 9/2 NADP+
+ 3H
2O
If the iron is initially in the +2 state, the reaction could be:
- Heme b2+ + 3O2 + 4 NADPH + 4 H+ → biliverdin + Fe2+ + CO + 4 NADP+ + 3H2O
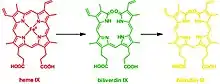
This reaction can occur in virtually every cell; the classic example is the formation of a contusion, which forms different chromogens as it gradually heals: (red) heme to (green) biliverdin to (yellow) bilirubin. In terms of molecular mechanisms, the enzyme facilitates the intramolecular hydroxylation of one meso carbon centre in the heme.[22]
Inducers
HO-1 is induced by countless molecules including heavy metals, statins, paclitaxel, rapamycin, probucol, nitric oxide, sildenafil, carbon monoxide, carbon monoxide-releasing molecules, and porphyrins.[23]
Phytochemical inducers of HO include: curcumin, resveratrol, piceatannol, caffeic acid phenethyl ester, dimethyl fumarate, fumaric acid esters, flavonoids, chalcones, ginkgo biloba, anthrocyanins, phlorotannins, carnosol, rosolic acid, and numerous other natural products.[23][24]
Endogenous inducers include i) lipids such as lipoxin and epoxyeicosatrienoic acid; and ii) peptides such as adrenomedullin and apolipoprotein; and iii) hemin.[23]
NRF2 inducers with downstream HO-1 induction include: genistein, 3-hydroxycoumarin, oleanolic acid, isoliquiritigenin, PEITC, diallyl trisulfide, oltipraz, benfotiamine, auranofin, acetaminophen, nimesulide, paraquat, ethoxyquin, diesel exhaust particles, silica, nanotubes, 15-deoxy-Δ12,14 prostaglandin J2, nitro-oleic acid, hydrogen peroxide, and succinylacetone.[25]
Roles in physiology
Heme oxygenase expression is induced by oxidative stress, and in animal models increasing this expression seems to be protective. Carbon monoxide released from heme oxygenase reactions can influence vascular tone independently or influence the function of nitric oxide synthase.
Endogenous carbon monoxide
The first detection of CO in humans was in 1949.[26] Sjöstrand determined that CO originated from the alpha-methene carbon of heme,[27] setting the stage for the molar ratio between hemin degradation and CO production to be established.[28] HO is the main source of endogenous CO production, though other minor contributors have been identified in recent years. CO is formed at a rate of 16.4 μmol/hour in the human body, ~86% originating from heme via heme oxygenase and ~14% from non-heme sources including: photooxidation, lipid peroxidation, and xenobiotics.[20] The average carboxyhemoglobin (CO-Hb) level in a non-smoker is between 0.2% and 0.85% CO-Hb (whereas a smoker may have between 4% and 10% CO-Hb),[29] though geographic location, occupation, health and behavior are contributing variables. Among these, the microbial HO system within the digestive tract is believed to contribute to systemic CO-Hb concentrations.[30]
References
- Kikuchi G, Yoshida T, Noguchi M (2005). "Heme oxygenase and heme degradation". Biochem. Biophys. Res. Commun. 338 (1): 558–567. doi:10.1016/j.bbrc.2005.08.020. PMID 16115609.
- Ryter SW, Alam J, Choi AM (2006). "Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications". Physiological Reviews. 86 (2): 583–650. doi:10.1152/physrev.00011.2005. PMID 16601269.
- Tenhunen R, Marver HS, Schmid R (1969). "Microsomal heme oxygenase. Characterization of the enzyme". The Journal of Biological Chemistry. 244 (23): 6388–6394. PMID 4390967.
- Motterlini R, Otterbein LE (2010). "The therapeutic potential of carbon monoxide". Nature Reviews. Drug Discovery. 9 (9): 728–743. doi:10.1038/nrd3228. PMID 20811383.
- Barton SG, Rampton DS, Winrow VR, Domizio P, Feakins RM (2003). "Expression of heat shock protein 32 (hemoxygenase-1) in the normal and inflamed human stomach and colon: an immunohistochemical study". Cell Stress & Chaperones. 8 (4): 329–334. doi:10.1379/1466-1268(2003)008<0329:eohsph>2.0.co;2. PMC 514904. PMID 15115285.
- Hopper CP, Meinel L, Steiger C, Otterbein LE (2018). "Where is the Clinical Breakthrough of Heme Oxygenase-1 / Carbon Monoxide Therapeutics?". Current Pharmaceutical Design. 24 (20): 2264–2282. doi:10.2174/1381612824666180723161811. PMID 30039755.
- Breman HJ, Wong RJ, Stevenson DK (2001). "Chapter 15: Sources, Sinks, and Measurement of Carbon Monoxide". In Wang R (ed.). Carbon Monoxide and Cardiovascular Functions (2nd ed.). CRC Press. ISBN 978-0-8493-1041-6.
- Lin, P (2008). "Ubiquitin–proteasome system mediates heme oxygenase-1 degradation through endoplasmic reticulum-associated degradation pathway". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1783 (10): 1826–1834. doi:10.1016/j.bbamcr.2008.05.008. PMID 18544348.
- Elbirt K, Bonkovsky H (1999). "Heme Oxygenase: Recent Advances in Understanding Its Regulation and Role". Proceedings of the Association of American Physicians. 111 (5): 438–447. doi:10.1111/paa.1999.111.5.438.
- https://medicalxpress.com/news/2018-11-enzyme-immune-cells-essential-role.html
- Uddin M, Jakaria M, Aleya L (2020). "Emerging promise of sulforaphane-mediated Nrf2 signaling cascade against neurological disorders". Science of the Total Environment. 707: 135624. doi:10.1016/j.scitotenv.2019.135624. PMID 31784171.
- Muñoz-Sánchez J, Chánez-Cárdenas ME (2014). "A review on hemeoxygenase-2: focus on cellular protection and oxygen response". Oxidative Medicine and Cellular Longevity. 2014 (1): 604981. doi:10.1155/2014/604981. PMC 4127239. PMID 25136403.
- Li C, Stocker R (2013). "Heme oxygenase and iron: from bacteria to humans". Redox Report. 14 (3): 95–101. doi:10.1179/135100009X392584. PMID 19490750.
- Protchenko O, Shakoury-Elizeh M, Keane P, Storey J, Androphy R, Philpott CC (2008). "Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae". Eukaryotic Cell. 7 (5): 859–871. doi:10.1128/EC.00414-07. PMC 2394968. PMID 18326586.
- Wilks A, Schmitt MP (1998). "Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle". The Journal of Biological Chemistry. 273 (2): 837–841. doi:10.1074/jbc.273.2.837. PMID 9422739.
- Maharshak N, Ryu HS, Fan TJ, Onyiah JC, Schulz S, Otterbein SL, Wong R, Hansen JJ, Otterbein LE, Carroll IM, Plevy SE (2015). "Escherichia coli heme oxygenase modulates host innate immune responses". Microbiology and Immunology. 59 (8): 452–465. doi:10.1111/1348-0421.12282. PMC 4582649. PMID 26146866.
- Frankenberg-Dinkel, Nicole (2004). "Bacterial Heme Oxygenases". Antioxidants & Redox Signaling. 6 (5): 825–834. doi:10.1089/ars.2004.6.825. PMID 15345142.
- LaMattina JW, Nix DB, Lanzilotta WN (2016). "Radical new paradigm for heme degradation in Escherichia coli O157:H7". Proceedings of the National Academy of Sciences of the United States of America. 113 (43): 12138–12143. doi:10.1073/pnas.1603209113. PMC 5087033. PMID 27791000.
- Matsui T, Nambu S, Ono Y, Goulding CW, Tsumoto K, Ikeda-Saito M (2013). "Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide". Biochemistry. 52 (18): 3025–3027. doi:10.1021/bi400382p. PMC 3672231. PMID 23600533.
- Wang R, ed. (2001). Carbon Monoxide and Cardiovascular Functions. CRC Press. p. 5. ISBN 978-1-4200-4101-9.
- Evans JP, Niemevz F, Buldain G, de Montellano PO (2008). "Isoporphyrin intermediate in heme oxygenase catalysis. Oxidation of alpha-meso-phenylheme". J. Biol. Chem. 283 (28): 19530–19539. doi:10.1074/jbc.M709685200. PMC 2443647. PMID 18487208. The reference does not give the exact stoichiometry of each reaction.
- Yoshida T, Taiko Migita C (2000). "Focused Review Mechanism of heme degradation by heme oxygenase". Journal of Inorganic Biochemistry. 82 (1–4): 33–41. doi:10.1016/S0162-0134(00)00156-2. PMID 11132636.
- Ferrándiz ML, Devesa I (2008). "Inducers of heme oxygenase-1". review. Current Pharmaceutical Design. 14 (5): 473–86. doi:10.2174/138161208783597399. PMID 18289074.
- Correa-Costa M, Otterbein LE (2014). "Eat to Heal: Natural Inducers of the Heme Oxygenase-1 System.". In Folkerts G, Garssen J (eds.). Pharma-Nutrition. secondary. AAPS Advances in the Pharmaceutical Sciences Series. 12. Springer, Cham. pp. 243–256. doi:10.1007/978-3-319-06151-1_12. ISBN 978-3-319-06150-4.
- Ma Q, He X (2012). "Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2". Pharmacol. Rev. 64 (4): 1055–1081. doi:10.1124/pr.110.004333. PMC 4648289. PMID 22966037.
- Sjöstrand T (1949). "Endogenous formation of carbon monoxide in man under normal and pathological conditions". Scandinavian Journal of Clinical and Laboratory Investigation. 1 (3): 201–214. doi:10.3109/00365514909069943.
- Sjöstrand T (1952). "The in vitro formation of carbon monoxide in blood". Acta Physiologica Scandinavica. 24 (4): 314–332. doi:10.1111/j.1748-1716.1952.tb00848.x. PMID 14952314.
- Coburn RF (1973). "Endogenous carbon monoxide metabolism". Annual Review of Medicine. 24: 241–250. doi:10.1146/annurev.me.24.020173.001325. PMID 4575855.
- Thom SR (2008). "Chapter 15: Carbon monoxide pathophysiology and treatment". In Neuman TS, Thom SR (eds.). Physiology and medicine of hyperbaric oxygen therapy. pp. 321–347. doi:10.1016/B978-1-4160-3406-3.50020-2. ISBN 9781416034063.
- Oleskin AV, Shenderov BA (2016). "Neuromodulatory effects and targets of the SCFAs and gasotransmitters produced by the human symbiotic microbiota". Microb. Ecol. Health Dis. 27: 30971. doi:10.3402/mehd.v27.30971. PMC 4937721. PMID 27389418.
External links
- Heme+Oxygenase at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 1.14.99.3