Hydrogenosome
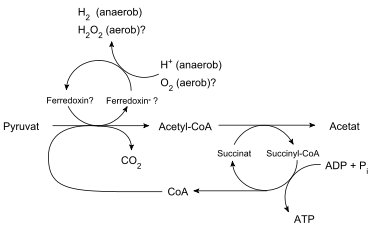
A hydrogenosome is a membrane-enclosed organelle of some anaerobic ciliates, trichomonads, fungi, and animals. The hydrogenosomes of trichomonads (the most studied of the hydrogenosome-containing microorganisms) produce molecular hydrogen, acetate, carbon dioxide and ATP by the combined actions of pyruvate:ferredoxin oxido-reductase, hydrogenase, acetate:succinate CoA transferase and succinate thiokinase. Superoxide dismutase, malate dehydrogenase (decarboxylating), ferredoxin, adenylate kinase and NADH:ferredoxin oxido-reductase are also localized in the hydrogenosome. It is nearly universally accepted that hydrogenosomes evolved from mitochondria.[2]
In 2010, scientists reported their discovery of the first known anaerobic metazoans with hydrogenosome-like organelles.[3]
History
Hydrogenosomes were isolated, purified, biochemically characterized and named in the early 1970s by D. G. Lindmark and M. Müller at Rockefeller University.[4] In addition to this seminal study on hydrogenosomes, they also demonstrated, for the first time, the presence of pyruvate:ferredoxin oxido-reductase and hydrogenase in eukaryotes. Further studies were subsequently conducted on the biochemical cytology and subcellular organization of anaerobic protozoan parasites (Trichomonas vaginalis, Tritrichomonas foetus, Monocercomonas sp., Giardia lamblia, Entamoeba sp., and Hexamita inflata). It is often forgotten that a Czechoslovakian group also biochemically described hydrogenosomes in 1973.[5]
Using information obtained from hydrogenosomal and biochemical cytology studies these researchers determined the mode of action of metronidazole (Flagyl) in 1976. Metronidazole is today recognized as the gold standard chemotherapeutic agent for the treatment of anaerobic infections caused by prokaryotes (Clostridium, Bacteroides, Helicobacter) and eukaryotes (Trichomonas, Tritrichomonas, Giardia, Entamoeba). Metronidazole is taken up by diffusion. Once taken up by anaerobes, it is non-enzymatically reduced by reduced ferredoxin which is produced by the action of pyruvate:ferredoxin oxido-reductase. This reduction creates products toxic to the anaerobic cell, and allows for selective accumulation of the drug in anaerobes.
Description
Hydrogenosomes are approximately 1 micrometre in diameter but under stress conditions can reach up to 2 micrometres[6] and are so called because they produce molecular hydrogen. Like mitochondria, they are bound by distinct double membranes and one has an inner membrane with some cristae-like projections.
Hydrogenosomes have evolved from mitochondria by loss of aerobiosis-related features in several lineages (not all hydrogenosomes are directly related).[7] In most cases, hydrogenosomes are genomeless, though genomes have persisted in some lineages such as Neocallimastix, Trichomonas vaginalis or Tritrichomonas foetus.[8] However, a hydrogenosomal genome has been detected in the cockroach ciliate Nyctotherus ovalis,[9] and the stramenopile Blastocystis.
The similarity between Nyctotherus and Blastocystis, which are only distantly related, is believed to be the result of convergent evolution, and calls into question whether there is a clear-cut distinction between mitochondria, hydrogenosomes, and mitosomes (another kind of degenerate mitochondria).[10]
Sources
The best studied hydrogenosomes are those of the sexually transmitted parasites Trichomonas vaginalis and Tritrichomonas foetus and those from rumen chytrids such as Neocallimastix.
The anaerobic ciliated protozoan Nyctotherus ovalis, found in the hindgut of several species of cockroach, has numerous hydrogenosomes that are intimately associated with endosymbiotic methane-producing archaea, the latter using the hydrogen produced by the hydrogenosomes. The matrix of N. ovalis hydrogenosomes contains ribosome-like particles of the same size as a numerous type of ribosome (70s) of the endosymbiotic methanogenic archaea. This suggested the presence of an organellar genome which was indeed discovered by Akhmanova and later partly sequenced by Boxma.[9][11]
Three multicellular species of Loricifera — Spinoloricus nov. sp., Rugiloricus nov. sp. and Pliciloricus nov. sp. — have been found deep in Mediterranean sediments, and use hydrogenosomes in their anaerobic metabolism cycle.[12][3]
In simple terms
One of the reasons that hydrogenosomes are of interest is because of the light they can cast on how life on earth may have evolved. Most life on earth is single celled, like Bacteria and Archaea. By contrast, plants and animals are made up of many cells. Crucially, each cell of a plant or animal is organised very differently from a bacterial cell. Amongst other things, animal and plant cells (which are special cases of what is known as 'eukaryotic' cells) contain mitochondria.
Mitochondria are like power stations for cells: They supply ATP molecules used throughout the cell as a power source by 'burning' carbohydrates and other fuels. They produce carbon dioxide and water as waste. To produce ATP, mitochondria require abundant supplies of oxygen.
Some single-celled eukaryotes live in places where oxygen is scarce or non-existent and use hydrogenosomes instead of mitochondria to produce ATP. Mitochondria do not function with little or no oxygen, but hydrogenosomes are able to produce ATP from most of the same fuels without using oxygen. In addition to ordinary carbon dioxide, the distinctive waste product of hydrogenosomes is hydrogen: The hydrogen they produce gives the organelles their name.
Hosting mitochondria appears to be a basal or near-basal trait among eukaryotes, the study of hydrogenosomes may shed light on how all multi-celled life developed. For example: Which came first: mitochondria, or hydrogenosomes, or some common ancestor?
See also
References
- Müller M, Lindmark DG (February 1978). "Respiration of hydrogenosomes of Tritrichomonas foetus. II. Effect of CoA on pyruvate oxidation". J. Biol. Chem. 253 (4): 1215–8. PMID 624726.
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM (12 Mar 2010). "Diversity and reductive evolution of mitochondria among microbial eukaryotes". Philos. Trans. R. Soc. Lond. B Biol. Sci. 365 (1541): 713–727. doi:10.1098/rstb.2009.0224. PMC 2817227. PMID 20124340.
- Danovaro R, Dell'anno A, Pusceddu A, Gambi C, Heiner I, Kristensen RM (April 2010). "The first metazoa living in permanently anoxic conditions". BMC Biol. 8 (1): 30. doi:10.1186/1741-7007-8-30. PMC 2907586. PMID 20370908.
- Lindmark DG, Müller M (November 1973). "Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism". J. Biol. Chem. 248 (22): 7724–8. PMID 4750424.
- Čerkasovová, A., Lukasová, G., Čerkasòv, J.. Kulda, J. (1973) Biochemical Characterization of Large Granule Fraction of Tritrichomonae foetus ( strain KV1 ) J. Protozool. 20, 525.
- Benchimol, M (2009). "Hydrogenosomes under microscopy". Tissue & Cell. 41 (3): 151–68. doi:10.1016/j.tice.2009.01.001. PMID 19297000.
- Embley TM. (2006) Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Philos Trans R Soc Lond B Biol Sci 361(1470):1055-67. Review.
- van der Giezen M, Tovar J, Clark CG (2005). "Mitochondrion-derived organelles in protists and fungi". Int. Rev. Cytol. International Review of Cytology. 244: 175–225. doi:10.1016/S0074-7696(05)44005-X. ISBN 9780123646484. PMID 16157181.
- Akhmanova A, Voncken F, van Alen T, et al. (December 1998). "A hydrogenosome with a genome" (PDF). Nature. 396 (6711): 527–8. doi:10.1038/25023. PMID 9859986.
- Stechmann, A; Hamblin, K; Pérez-brocal, V; Gaston, D; Richmond, Gs; Van, Der; Clark, Cg; Roger, Aj (Apr 2008). "Organelles in Blastocystis that Blur the Distinction between Mitochondria and Hydrogenosomes". Current Biology. 18 (8): 580–5. doi:10.1016/j.cub.2008.03.037. PMC 2428068. PMID 18403202.
- Boxma B, de Graaf RM, van der Staay GW, et al. (March 2005). "An anaerobic mitochondrion that produces hydrogen". Nature. 434 (7029): 74–9. doi:10.1038/nature03343. PMID 15744302.
- Fang J (April 6, 2010). "Animals thrive without oxygen at sea bottom". Nature. Nature Publishing Group. 464 (7290): 825. doi:10.1038/464825b. PMID 20376121.
