GroEL
GroEL is a protein which belongs to the chaperonin family of molecular chaperones, and is found in many bacteria.[5] It is required for the proper folding of many proteins. To function properly, GroEL requires the lid-like cochaperonin protein complex GroES. In eukaryotes the organellar proteins Hsp60 and Hsp10 are structurally and functionally nearly identical to GroEL and GroES, respectively, due to their endosymbiotic origin.
HSP60 is implicated in mitochondrial protein import and macromolecular assembly. It may facilitate the correct folding of imported proteins, and may also prevent misfolding and promote the refolding and proper assembly of unfolded polypeptides generated under stress conditions in the mitochondrial matrix. HSP60 interacts with HRAS and with HBV protein X and HTLV-1 protein p40tax. HSP60 belongs to the chaperonin (HSP60) family. Note: This description may include information from UniProtKB.
Alternate Names: 60 kDa chaperonin, Chaperonin 60, CPN60, Heat shock protein 60, HSP-60, HuCHA60, Mitochondrial matrix protein P1, P60 lymphocyte protein, HSPD1
Heat shock protein 60 (HSP60) is a mitochondrial chaperonin that is typically held responsible for the transportation and refolding of proteins from the cytoplasm into the mitochondrial matrix. In addition to its role as a heat shock protein, HSP60 functions as a chaperonin to assist in folding linear amino acid chains into their respective three-dimensional structure. Through the extensive study of groEL, HSP60’s bacterial homolog, HSP60 has been deemed essential in the synthesis and transportation of essential mitochondrial proteins from the cell's cytoplasm into the mitochondrial matrix. Further studies have linked HSP60 to diabetes, stress response, cancer and certain types of immunological disorders.
Discovery
Not much is known about the function of HSP60. Mammalian HSP60 was first reported as a mitochondrial P1 protein. It was subsequently cloned and sequenced by Radhey Gupta and coworkers.[6] The amino acid sequence showed a strong homology to GroEL. It was initially believed that HSP60 functioned only in the mitochondria and that there was no equivalent protein located in the cytoplasm. Recent discoveries have discredited this claim and have suggested that there is a recognizable difference between HSP60 in the mitochondria and in the cytoplasm.[7] A similar protein structure exists in the chloroplast of certain plants. This protein presence provides evidence for the evolutionary relationship of the development of the mitochondria and the chloroplast by means of endosymbiosis.[6]
Structure
Under normal physiological conditions, HSP60 is a 60 kilodalton oligomer composed of monomers that form a complex arranged as two stacked heptameric rings.[8] This double ring structure forms a large central cavity in which the unfolded protein binds via hydrophobic interactions.[9] This structure is typically in equilibrium with each of its individual components: monomers, heptamers, and tetradeceamers.[10] Recent studies have begun to suggest that in addition to its typical location in the mitochondria, HSP60 can also be found in the cytoplasm under normal physiological conditions.[7]
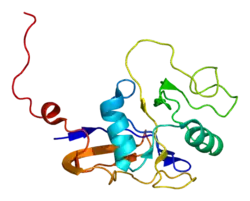
Each subunit of HSP60 has three domains: the apical domain, the equatorial domain, and the intermediate domain.[11] The equatorial domain contains the binding site for ATP and for the other heptameric ring. The intermediate domain binds the equatorial domain and the apical domain together.[11] The intermediate domain induces a conformational change when ATP is bound allowing for an alternation between the hydrophilic and hydrophobic substrate binding sites.[11] In its inactive state, the protein is in a hydrophobic state. When activated by ATP, the intermediate domain undergoes a conformational change that exposes the hydrophilic region. This insures fidelity in protein binding.[11] Chaperonin 10 aids HSP60 in folding by acting as a dome-like cover on the ATP active form of HSP60. This causes the central cavity to enlarge and aids in protein folding.[11] See the above figure for further detail on the structure.
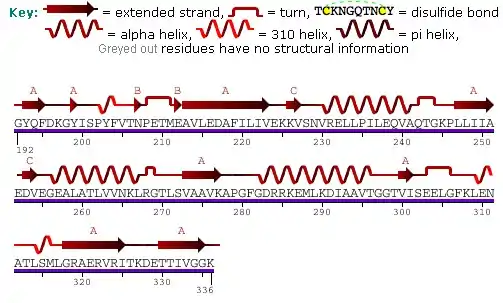
The mitochondrial HSP60 sequence contains a series of G repeats at the C-terminal.[6] The structure and function of this sequence is not quite known. The N-terminal contains a pre-sequence of hydroxylated amino acids, namely arginine, lysine, serine, and threonine, which serve as directors for the importation of the protein into the mitochondria.[6]
The predicted structure of HSP60 includes several vertical sine waves, alpha helices, beta sheets, and 90 degree turns. There are regions of hydrophobicity where the protein presumably spans the membrane. There are also three N-linked glycosylation sites at positions 104, 230, 436.[9] The sequence and secondary structure for the mitochondrial protein are illustrated in the above image obtained from the Protein Data Bank.
Newer information has begun to suggest that the HSP60 found in the mitochondria differs from that of the cytoplasm. With respect to the amino acid sequence, the cytoplasmic HSP60 has an N-terminal sequence not found in the mitochondrial protein.[7] In gel electrophoresis analysis, significant differences were found in the migration of cytoplasmic and mitochondrial HSP60. The cytoplasmic HSP60 contains a signal sequence of 26 amino acids on the N terminus. This sequence is highly degenerate and is capable of folding into amphiphilic helix.[7] Antibodies against HSP60 targeted both the mitochondrial and cytoplasmic form.[7] Nonetheless, antibodies against the signal sequence targeted only the cytoplasmic form. Under normal physiological condition, both are found in relatively equal concentrations.[7] In times of stress or high need of HSP60 in either the cytoplasm or the mitochondria, the cell is capable for compensating by increasing the presence of HSP60 in one compartment and decreasing its concentration in the opposite compartment.
Function
Common
Heat shock proteins are amongst the most evolutionarily conserved of proteins.[10] The significant function, structural, and sequential homology between HSP60 and its prokaryotic homolog, groEL, demonstrates this level of conservation. Moreover, HSP60’s amino acid sequence bears a similarity to its homolog in plants, bacteria, and humans.[13] Heat shock proteins are primarily responsible for maintaining the integrity of cellular proteins particularly in response to environmental changes. Stresses such as temperature, concentration imbalance, pH change, and toxins can all induce heat shock proteins to maintain the conformation of the cell’s proteins. HSP60 aids in the folding and conformation maintenance of approximately 15-30% of all cellular proteins.[11] In addition to HSP60’s typical role as a heat shock protein, studies have shown that HSP60 plays an important role in the transport and maintenance of mitochondrial proteins as well as the transmission and replication of mitochondrial DNA.
Mitochondrial protein transport
HSP60 possesses two main responsibilities with respect to mitochondrial protein transport. It functions to catalyze the folding of proteins destined for the matrix and maintains protein in an unfolded state for transport across the inner membrane of the mitochondria.[14] Many proteins are targeted for processing in the matrix of the mitochondria but then are quickly exported to other parts of the cell. The hydrophobic portion HSP60 is responsible for maintaining the unfolded conformation of the protein for transmembrane transport.[14] Studies have shown how HSP60 binds to incoming proteins and induces conformational and structural changes. Subsequent changes in ATP concentrations hydrolyze the bonds between the protein and HSP60 which signals the protein to exit the mitochondria.[14] HSP60 is also capable of distinguishing between proteins designated for export and proteins destined to remain in the mitochondrial matrix by looking for an amphiphilic alpha-helix of 15-20 residues.[14] The existence of this sequence signals that the protein is to be exported while the absence signals that the protein is to remain in the mitochondria. The precise mechanism is not yet entirely understood.
DNA metabolism
In addition to its critical role in protein folding, HSP60 is involved in the replication and transmission of mitochondrial DNA. In extensive studies of HSP60 activity in Saccharomyces cerevisiae, scientists have proposed that HSP60 binds preferentially to the single stranded template DNA strand in a tetradecamer like complex [15] This tetradecamer complex interacts with other transcriptional elements to serve as a regulatory mechanism for the replication and transmission of mitochondrial DNA. Mutagenic studies have further supported HSP60 regulatory involvement in the replication and transmission of mitochondrial DNA.[16]Mutations in HSP60 increase the levels of mitochondrial DNA and result in subsequent transmission defects.
Cytoplasmic vs mitochondrial HSP60
In addition to the already illustrated structural differences between cytoplasmic and mitochondrial HSP60, there are marked functional differences. Studies have suggested that HSP60 plays a key role in preventing apoptosis in the cytoplasm. The cytoplasmic HSP60 forms a complex with proteins responsible for apoptosis and regulates the activity of these proteins.[7] The cytoplasmic version is also involved in immune response and cancer.[7] These two aspects will be elaborated on later. Extremely recent investigations have begun to suggest a regulatory correlation between HSP60 and the glycolytic enzyme, 6-phosphofructokinase-1. Although not much information is available, cytoplasmic HSP60 concentrations have influenced the expression of 6-phosphofructokinase in glycolysis.[17] Despite these marked differences between the cytoplasmic and mitochondrial form, experimental analysis has shown that the cell is quickly capable of moving cytoplasmic HSP60 into the mitochondria if environmental conditions demand a higher presence of mitochondrial HSP60.[7]
Synthesis and assembly
HSP60 is typically found in the mitochondria and has been found in organelles of endosymbiotic origin. HSP60 monomers form two heptameric rings that bind to the surface of linear proteins and catalyze their folding in an ATP dependent process.[18] HSP60 subunits are encoded by nuclear genes and translated into the cytosol. These subunits then move into the mitochondria where they are processed by other HSP60 molecules.[9] Several studies have shown how HSP60 proteins must be present in the mitochondria for the synthesis and assembly of additional HSP60 components.[9] There is a direct positive correlation between the presence of HSP60 proteins in the mitochondria and the production of additional HSP60 protein complexes.
The kinetics of assembly of HSP60 subunits into the 2-heptameric rings takes two minutes. The subsequent protease-resistant HSP60 is formed in a half-time of 5–10 minutes.[9] This rapid synthesis indicates that there is an ATP-dependent interaction where the formed HSP60 complex stabilizes the intermediate of the HSP60 assembly complex, effectively serving as a catalyst.[9] The necessity of preexisting HSP60 in order to synthesize additional HSP60 molecules supports the endosymbiotic theory of the origin of mitochondria. There must have been a rudimentary prokaryotic homologous protein that was capable of similar self-assembly.
Immunological role
As discussed above, HSP60 has generally been known as a chaperonin which assists in protein folding in mitochondria. However, some new research has indicated that HSP60 possibly plays a role in a “danger signal cascade” immune response.[19] There is also mounting evidence that it plays a role in autoimmune disease.
Infection and disease are extremely stressful on the cell. When a cell is under stress, it naturally increases the production of stress proteins, including heat shock proteins such as HSP60. In order for HSP60 to act as a signal it must be present in the extracellular environment. In recent research “it has emerged that…chaperonin 60 can be found on the surface of various prokaryotic and eukaryotic cells, and can even be released from cells”.[11] According to recent research, many different types of heat shock proteins are used in immune response signaling, but it appears that different proteins act and respond differently to other signaling molecules. HSP60 has been shown to be released from specific cells like peripheral blood mononuclear cells (PBMCs) when there are lipopolysaccharides (LPS) or GroEL present. This suggests that the cell has different receptors and responses to human and bacterial HSP60.[19] In addition, it has been shown that HSP60 has the capability “of activating monocytes, macrophages and dendritic cells…and also of inducing secretion of a wide range of cytokines.” [19] The fact that HSP60 responds to other signal molecules like LPS or GroEL and has the ability to activate certain types of cells supports the idea that HSP60 is part of a danger signal cascade which is involved in activating an immune response.
There is however, a twist in the immunological role of HSP60. As mentioned above, there are two different types of HSP60 proteins, bacterial as well as mammalian. Since they are very similar in sequence, bacterial HSP60 wouldn’t be expected to cause a large immune response in humans. The immune system is “designed to ignore ‘self’, that is, host constituents; however, paradoxically, this is not the case with chaperonins”.[11] It has been found that many anti-chaperonin antibodies exist and are associated with many autoimmune diseases. According to Ranford, et al. experiments have been performed which have shown that antibodies which are “generated by a human host after exposure to bacterial chaperonin 60 proteins” can cross-react with human chaperonin 60 proteins.[11] Bacterial HSP60 is causing the immune system to create anti-chaperonin antibodies, even though bacterial and human HSP60 have similar protein sequences. These new antibodies are then recognizing and attacking human HSP60 which causes an autoimmune disease. This suggests that HSP60 may play a role in autoimmunity, however more research needs to be done in order to discover more completely its role in this disease.
Stress response
HSP60, as a mitochondrial protein, has been shown to be involved in stress response as well. The heat shock response is a homeostatic mechanism that protects a cell from damage by upregulating the expression of genes that code for HSP60.[20] The upregulation of HSP60 production allows for the maintenance of other cellular processes occurring in the cell, especially during stressful times. In one experiment, investigators treated various mice with L-DOPA and discovered significant upregulation of HSP60 expression in the mitochondria and HSP70 expression in the cytoplasm. Researchers concluded that the heat shock signal pathway serves as “the basic mechanism of defense against neurotoxicity elicited by free radical oxygen and nitrogen species produced in aging and neurodegenerative disorders”.[21] Several studies have shown that HSP60 and other heat shock proteins are necessary for cellular survival under toxic or stressful circumstances.[22]
Relationship to cancer
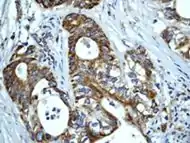
Human Hsp60, the product of the HSPD1 gene, is a Group I mitochondrial chaperonin, phylogenetically related to bacterial GroEL. Recently, the presence of Hsp60 outside the mitochondria and outside the cell, e.g. in circulating blood, has been reported [1], [2]. Although it is assumed that Hsp60 extra-mitochondrial molecule is identical to the mitochondrial one, this has not yet been fully elucidated. Despite the increasing amount of experimental evidences showing Hsp60 outside the cell, it is not yet clear how general this process is and what are the mechanisms responsible for Hsp60 translocation outside the cell. Neither of these questions has been definitively answered, whereas there is some information regarding extracellular Hsp70. This chaperone was also classically regarded as an intracellular protein like Hsp60, but in the last few years considerable evidences showed its pericellular and extracellular residence
HSP60 has been shown to influence apoptosis in tumor cells which seems to be associated with a change in expression levels. There is some inconsistency in that some research shows a positive expression while other research shows a negative expression, and it seems to depend on the type of cancer. There are different hypotheses to explain the effects of positive versus negative expression. Positive expression seems to inhibit “apoptotic and necrotic cell death” while negative expression is thought to play a part “in activation of apoptosis”.[23][24]
As well as influencing apoptosis, HSP60 changes in expression level have been shown to be “useful new biomarkers for diagnostic and prognostic purposes.” [23] According to Lebret et al., a loss of HSP60 expression “indicates a poor prognosis and the risk of developing tumor infiltration” specifically with bladder carcinomas, but that does not necessarily hold true for other types of cancers.[25] For example, ovarian tumors research has shown that over expression is correlated with a better prognosis while a decreased expression is correlated with an aggressive tumor.[25] All this research indicates that it may be possible for HSP60 expression to be used in predicting survival for certain types of cancer and therefore may be able to identify patients who could benefit from certain treatments.[24]
Mechanism
Within the cell, the process of GroEL/ES mediated protein folding involves multiple rounds of binding, encapsulation, and release of substrate protein. Unfolded substrate proteins bind to a hydrophobic binding patch on the interior rim of the open cavity of GroEL, forming a binary complex with the chaperonin. Binding of substrate protein in this manner, in addition to binding of ATP, induces a conformational change that allows association of the binary complex with a separate lid structure, GroES. Binding of GroES to the open cavity of the chaperonin induces the individual subunits of the chaperonin to rotate such that the hydrophobic substrate binding site is removed from the interior of the cavity, causing the substrate protein to be ejected from the rim into the now largely hydrophilic chamber. The hydrophilic environment of the chamber favors the burying of hydrophobic residues of the substrate, inducing substrate folding. Hydrolysis of ATP and binding of a new substrate protein to the opposite cavity sends an allosteric signal causing GroES and the encapsulated protein to be released into the cytosol. A given protein will undergo multiple rounds of folding, returning each time to its original unfolded state, until the native conformation or an intermediate structure committed to reaching the native state is achieved. Alternatively, the substrate may succumb to a competing reaction, such as misfolding and aggregation with other misfolded proteins.[26]
Thermodynamics
The constricted nature of the interior of the molecular complex strongly favors compact molecular conformations of the substrate protein. Free in solution, long-range, non-polar interactions can only occur at a high cost in entropy. In the close quarters of the GroEL complex, the relative loss of entropy is much smaller. The method of capture also tends to concentrate the non-polar binding sites separately from the polar sites. When the GroEL non-polar surfaces are removed, the chance that any given non-polar group will encounter a non-polar intramolecular site are much greater than in bulk solution. The hydrophobic sites which were on the outside are gathered together at the top of the cis domain and bind each other. The geometry of GroEL requires that the polar structures lead, and they envelop the non-polar core as it emerges from the trans side.
Structure
Structurally, GroEL is a dual-ringed tetradecamer, with both the cis and trans rings consisting of seven subunits each. The conformational changes that occur within the central cavity of GroEL cause for the inside of GroEL to become hydrophilic, rather than hydrophobic, and is likely what facilitates protein folding.
 GroEL (side)
GroEL (side) GroEL (top)
GroEL (top)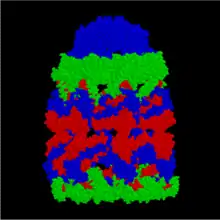 GroES/GroEL complex (side)
GroES/GroEL complex (side)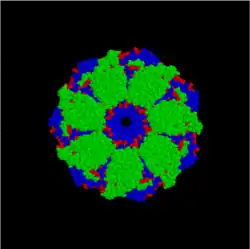 GroES/GroEL complex (top)
GroES/GroEL complex (top)
The key to the activity of GroEL is in the structure of the monomer. The Hsp60 monomer has three distinct sections separated by two hinge regions. The apical section contains many hydrophobic binding sites for unfolded protein substrates. Many globular proteins won't bind to the apical domain because their hydrophobic parts are clustered inside, away from the aqueous medium since this is the thermodynamically optimal conformation. Thus, these "substrate sites" will only bind to proteins which are not optimally folded. The apical domain also has binding sites for the Hsp10 monomers of GroES.
The equatorial domain has a slot near the hinge point for binding ATP, as well as two attachment points for the other half of the GroEL molecule. The rest of the equatorial section is moderately hydrophilic.
The addition of ATP and GroES has a drastic effect on the conformation of the cis domain. This effect is caused by flexion and rotation at the two hinge points on the Hsp60 monomers. The intermediate domain folds down and inward about 25° on the lower hinge. This effect, multiplied through the cooperative flexing of all monomers, increases the equatorial diameter of the GroEL cage. But the apical domain rotates a full 60° up and out on the upper hinge, and also rotates 90° around the hinge axis. This motion opens the cage very widely at the top of the cis domain, but completely removes the substrate binding sites from the inside of the cage.
Interactions
GroEL has been shown to interact with GroES,[27][28] ALDH2,[28] Caspase 3[27][29] and Dihydrofolate reductase.[30]
Phage T4 morphogenesis
The genes of bacteriophage (phage) T4 that encode proteins with a role in determining phage T4 structure were identified using conditional lethal mutants.[31] Most of these proteins proved to be either major or minor structural components of the completed phage particle. However among the gene products (gps) necessary for phage assembly, Snustad[32] identified a group of gps that act catalytically rather than being incorporated themselves into the phage structure. These catalytic gps included gp31. The bacterium E. coli is the host for phage T4, and the phage encoded gp31 protein appears to be functionally homologous to E. coli chaparone protein GroES and able to substitute for it in the assembly of phage T4 virions during infection.[5] The role of the phage encoded gp31 protein appears be to interact with the E. coli host encoded GroEL protein to assist in the correct folding and assembly of the major phage head capsid protein of the phage, gp23.[5]
See also
References
- GRCh38: Ensembl release 89: ENSG00000144381 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000025980 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Zeilstra-Ryalls J, Fayet O, Georgopoulos C (1991). "The universally conserved GroE (Hsp60) chaperonins". Annu. Rev. Microbiol. 45: 301–25. doi:10.1146/annurev.mi.45.100191.001505. PMID 1683763.
- Gupta RS (January 1995). "Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells". Mol. Microbiol. 15 (1): 1–11. doi:10.1111/j.1365-2958.1995.tb02216.x. PMID 7752884.
- Itoh H, Komatsuda A, Ohtani H, et al. (December 2002). "Mammalian HSP60 is quickly sorted into the mitochondria under conditions of dehydration". Eur. J. Biochem. 269 (23): 5931–8. doi:10.1046/j.1432-1033.2002.03317.x. PMID 12444982.
- Cheng MY, Hartl FU, Horwich AL (November 1990). "The mitochondrial chaperonin hsp60 is required for its own assembly". Nature. 348 (6300): 455–8. doi:10.1038/348455a0. PMID 1978929.
- Fenton WA, et al. (October 1994). "Residues in chaperonin GroEL required for polypeptide binding and release". Nature. 371 (6498): 614–9. doi:10.1038/371614a0. PMID 7935796.
- Habich C, et al. (March 2007). "Heat shock protein 60: regulatory role on innate immune cells". Cell. Mol. Life Sci. 64 (6): 742–51. doi:10.1007/s00018-007-6413-7. PMID 17221165.
- Ranford JC, et al. (September 2000). "Chaperonins are cell-signalling proteins: the unfolding biology of molecular chaperones". Expert Rev Mol Med. 2 (8): 1–17. doi:10.1017/S1462399400002015. PMID 14585136.
- PDB: 1SRV; Walsh MA, et al. (June 1999). "Taking MAD to the extreme: ultrafast protein structure determination". Acta Crystallogr. D. 55 (6): 1168–73. doi:10.1107/S0907444999003698. PMID 10329779.
- Johnson RB, et al. (2003). "Cloning and characterization of the yeast chaperonin HSP60 gene". Genetics. 84 (2): 295–300. doi:10.1016/0378-1119(89)90503-9. PMID 2575559.
- Koll H, et al. (March 1992). "Antifolding activity of hsp60 couples protein import into the mitochondrial matrix with export to the intermembrane space" (PDF). Cell. 68 (6): 1163–75. doi:10.1016/0092-8674(92)90086-R. PMID 1347713.
- Kaufman, BA. Studies on mitochondria DNA nucleoids in Saccharomyces cerevisia: identification of bifunctional proteins. In Genetics and Development, UT Southwestern Medical Center at Dallas, Dallas, TX. 241pp.
- Kaufman, B. A. (2003). "A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae". The Journal of Cell Biology. 163 (3): 457–461. doi:10.1083/jcb.200306132. ISSN 0021-9525. PMC 2173642. PMID 14597775.
- Koll H, et al. (1992). "Antifolding Activity of HSP60 Couples Protein Import into the Mitochondrial Matrix with Export to the Intermembrane Space" (PDF). Cell. 68 (6): 1163–75. doi:10.1016/0092-8674(92)90086-R. PMID 1347713.
- Itoh H, et al. (December 2002). "Mammalian HSP60 is quickly sorted into the mitochondria under conditions of dehydration". Eur. J. Biochem. 269 (23): 5931–8. doi:10.1046/j.1432-1033.2002.03317.x. PMID 12444982.
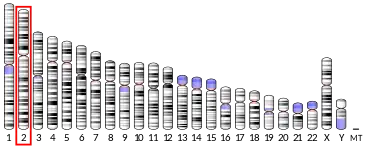
- Hansen JJ, Bross P, Westergaard M, et al. (January 2003). "Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter". Hum. Genet. 112 (1): 71–7. doi:10.1007/s00439-002-0837-9. PMID 12483302.
- Vargas-Parada L, Solis C (2001). "Heat Shock and stress response of Taenia solium and T. crassiceps". Parasitology. 122 (5): 583–8. doi:10.1017/s0031182001007764.
- Calabrese V, Mancuso C, Ravagna A, et al. (May 2007). "In vivo induction of heat shock proteins in the substantia nigra following L-DOPA administration is associated with increased activity of mitochondrial complex I and nitrosative stress in rats: regulation by glutathione redox state". J. Neurochem. 101 (3): 709–17. doi:10.1111/j.1471-4159.2006.04367.x. PMID 17241115.
- Rossi MR, Somji S, Garrett SH, Sens MA, Nath J, Sens DA (December 2002). "Expression of hsp 27, hsp 60, hsc 70, and hsp 70 stress response genes in cultured human urothelial cells (UROtsa) exposed to lethal and sublethal concentrations of sodium arsenite". Environ. Health Perspect. 110 (12): 1225–32. doi:10.1289/ehp.021101225. PMC 1241110. PMID 12460802.
- Cappello F, Di Stefano A, David S, et al. (November 2006). "Hsp60 and Hsp10 down-regulation predicts bronchial epithelial carcinogenesis in smokers with chronic obstructive pulmonary disease". Cancer. 107 (10): 2417–24. doi:10.1002/cncr.22265. PMID 17048249.
- Urushibara M, Kageyama Y, Akashi T, et al. (January 2007). "HSP60 may predict good pathological response to neoadjuvant chemoradiotherapy in bladder cancer". Jpn. J. Clin. Oncol. 37 (1): 56–61. doi:10.1093/jjco/hyl121. PMID 17095522.
- Lebret T, Watson RW, Molinié V, et al. (September 2003). "Heat shock proteins HSP27, HSP60, HSP70, and HSP90: expression in bladder carcinoma". Cancer. 98 (5): 970–7. doi:10.1002/cncr.11594. PMID 12942564.
- Horwich AL, Fenton WA, Chapman E, Farr GW (2007). "Two families of chaperonin: physiology and mechanism". Annu. Rev. Cell Dev. Biol. 23: 115–45. doi:10.1146/annurev.cellbio.23.090506.123555. PMID 17489689.
- Samali A, Cai J, Zhivotovsky B, Jones DP, Orrenius S (Apr 1999). "Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells". EMBO J. 18 (8): 2040–8. doi:10.1093/emboj/18.8.2040. PMC 1171288. PMID 10205158.
- Lee KH, Kim HS, Jeong HS, Lee YS (Oct 2002). "Chaperonin GroESL mediates the protein folding of human liver mitochondrial aldehyde dehydrogenase in Escherichia coli". Biochem. Biophys. Res. Commun. 298 (2): 216–24. doi:10.1016/S0006-291X(02)02423-3. PMID 12387818.
- Xanthoudakis S, Roy S, Rasper D, Hennessey T, Aubin Y, Cassady R, Tawa P, Ruel R, Rosen A, Nicholson DW (Apr 1999). "Hsp60 accelerates the maturation of pro-caspase-3 by upstream activator proteases during apoptosis". EMBO J. 18 (8): 2049–56. doi:10.1093/emboj/18.8.2049. PMC 1171289. PMID 10205159.
- Mayhew M, da Silva AC, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU (Feb 1996). "Protein folding in the central cavity of the GroEL-GroES chaperonin complex". Nature. 379 (6564): 420–6. doi:10.1038/379420a0. PMID 8559246. S2CID 4310511.
- Edgar RS, Epstein RH (February 1965). "The genetics of a bacterial virus". Scientific American. 212 (2): 70–8. Bibcode:1965SciAm.212b..70E. doi:10.1038/scientificamerican0265-70. PMID 14272117.
- Snustad DP (August 1968). "Dominance interactions in Escherichia coli cells mixedly infected with bacteriophage T4D wild-type and amber mutants and their possible implications as to type of gene-product function: catalytic vs. stoichiometric". Virology. 35 (4): 550–63. doi:10.1016/0042-6822(68)90285-7. PMID 4878023.
Further reading
- Tabibzadeh S, Broome J (1999). "Heat shock proteins in human endometrium throughout the menstrual cycle". Infect Dis Obstet Gynecol. 7 (1–2): 5–9. doi:10.1002/(SICI)1098-0997(1999)7:1/2<5::AID-IDOG2>3.0.CO;2-Y. PMC 1784709. PMID 10231001.
- Schäfer C, Williams JA (2000). "Stress kinases and heat shock proteins in the pancreas: possible roles in normal function and disease". J. Gastroenterol. 35 (1): 1–9. doi:10.1080/003655200750024443. hdl:2027.42/42441. PMID 10632533. S2CID 9706591.
- Moseley P (2000). "Stress proteins and the immune response". Immunopharmacology. 48 (3): 299–302. doi:10.1016/S0162-3109(00)00227-7. PMID 10960671.
- Liu Y, Steinacker JM (2001). "Changes in skeletal muscle heat shock proteins: pathological significance". Front. Biosci. 6: D12-25. doi:10.2741/Liu. PMID 11145923.
- Van Maele B, Debyser Z (2005). "HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins". AIDS Rev. 7 (1): 26–43. PMID 15875659.
- Hochstrasser DF, Frutiger S, Paquet N, Bairoch A, Ravier F, Pasquali C, Sanchez JC, Tissot JD, Bjellqvist B, Vargas R (1992). "Human liver protein map: a reference database established by microsequencing and gel comparison". Electrophoresis. 13 (12): 992–1001. doi:10.1002/elps.11501301201. PMID 1286669. S2CID 23518983.
- Ikawa S, Weinberg RA (1992). "An interaction between p21ras and heat shock protein hsp60, a chaperonin". Proc. Natl. Acad. Sci. U.S.A. 89 (6): 2012–6. doi:10.1073/pnas.89.6.2012. PMC 48586. PMID 1347942.
- Brudzynski K, Martinez V, Gupta RS (1992). "Immunocytochemical localization of heat-shock protein 60-related protein in beta-cell secretory granules and its altered distribution in non-obese diabetic mice". Diabetologia. 35 (4): 316–24. doi:10.1007/BF00401198. PMID 1516759.
- Dawson SJ, White LA (1992). "Treatment of Haemophilus aphrophilus endocarditis with ciprofloxacin". J. Infect. 24 (3): 317–20. doi:10.1016/S0163-4453(05)80037-4. PMID 1602151.
- Singh B, Patel HV, Ridley RG, Freeman KB, Gupta RS (1990). "Mitochondrial import of the human chaperonin (HSP60) protein". Biochem. Biophys. Res. Commun. 169 (2): 391–6. doi:10.1016/0006-291X(90)90344-M. PMID 1972619.
- Venner TJ, Singh B, Gupta RS (1990). "Nucleotide sequences and novel structural features of human and Chinese hamster hsp60 (chaperonin) gene families". DNA Cell Biol. 9 (8): 545–52. doi:10.1089/dna.1990.9.545. PMID 1980192.
- Ward LD, Hong J, Whitehead RH, Simpson RJ (1990). "Development of a database of amino acid sequences for human colon carcinoma proteins separated by two-dimensional polyacrylamide gel electrophoresis". Electrophoresis. 11 (10): 883–91. doi:10.1002/elps.1150111019. PMID 2079031. S2CID 21541503.
- Jindal S, Dudani AK, Singh B, Harley CB, Gupta RS (1989). "Primary structure of a human mitochondrial protein homologous to the bacterial and plant chaperonins and to the 65-kilodalton mycobacterial antigen". Mol. Cell. Biol. 9 (5): 2279–83. doi:10.1128/mcb.9.5.2279. PMC 363030. PMID 2568584.
- Waldinger D, Eckerskorn C, Lottspeich F, Cleve H (1988). "Amino-acid sequence homology of a polymorphic cellular protein from human lymphocytes and the chaperonins from Escherichia coli (groEL) and chloroplasts (Rubisco-binding protein)". Biol. Chem. Hoppe-Seyler. 369 (10): 1185–9. doi:10.1515/bchm3.1988.369.2.1185. PMID 2907406.
- Kreisel W, Hildebrandt H, Schiltz E, Köhler G, Spamer C, Dietz C, Mössner W, Heilmann C (1994). "Immuno-gold electron microscopical detection of heat shock protein 60 (hsp60) in mitochondria of rat hepatocytes and myocardiocytes". Acta Histochem. 96 (1): 51–62. doi:10.1016/s0065-1281(11)80009-7. PMID 7518175.
- Corbett JM, Wheeler CH, Baker CS, Yacoub MH, Dunn MJ (1994). "The human myocardial two-dimensional gel protein database: update 1994". Electrophoresis. 15 (11): 1459–65. doi:10.1002/elps.11501501209. PMID 7895732. S2CID 33359306.
- Baca-Estrada ME, Gupta RS, Stead RH, Croitoru K (1994). "Intestinal expression and cellular immune responses to human heat-shock protein 60 in Crohn's disease". Dig. Dis. Sci. 39 (3): 498–506. doi:10.1007/BF02088334. PMID 7907543. S2CID 22032288.
- Vélez-Granell CS, Arias AE, Torres-Ruíz JA, Bendayan M (1994). "Molecular chaperones in pancreatic tissue: the presence of cpn10, cpn60 and hsp70 in distinct compartments along the secretory pathway of the acinar cells". J. Cell Sci. 107 (3): 539–49. PMID 7911805.
- Mayhew M, da Silva AC, Martin J, Erdjument-Bromage H, Tempst P, Hartl FU (1996). "Protein folding in the central cavity of the GroEL-GroES chaperonin complex". Nature. 379 (6564): 420–6. doi:10.1038/379420a0. PMID 8559246. S2CID 4310511.
- Tabibzadeh S, Kong QF, Satyaswaroop PG, Babaknia A (1996). "Heat shock proteins in human endometrium throughout the menstrual cycle". Hum. Reprod. 11 (3): 633–40. doi:10.1093/humrep/11.3.633. PMID 8671282.
External links
- GroEL+Protein at the US National Library of Medicine Medical Subject Headings (MeSH)
- "Palaeos Bacteria: Pieces: GroEL". Archived from the original on 2007-04-26. (No rights reserved)
- 3D macromolecular structures of GroEL in EMDB