Prokaryotic ubiquitin-like protein
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote Mycobacterium tuberculosis.[1] Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation. However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase.
| Pup-like protein family | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
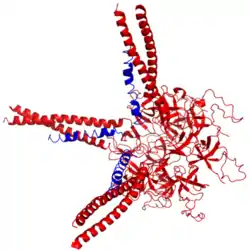 Three Prokaryotic ubiquitin-like proteins (blue) bound to proteasomal ATPase Mpa (red | |||||||||||
| Identifiers | |||||||||||
| Symbol | Pup/Mpa | ||||||||||
| Pfam | PF05639 | ||||||||||
| InterPro | IPR008515 | ||||||||||
| |||||||||||
Similar to ubiquitin, Pup is attached to specific lysine residues of substrate proteins by isopeptide bonds; this is called pupylation. It is then recognized by the protein Mycobacterium proteasomal ATPase (Mpa), in a mechanism that induces folding of Pup.[2] Mpa delivers the substrate protein to the proteasome for degradation by coupling of ATP hydrolysis.
The discovery of Pup indicates that like eukaryotes, bacteria may use a small-protein modifier to control protein stability.
The Pup gene encodes a 64–amino acid protein with a molecular size of about 6.9 kDa.[3]
Pup is an intrinsically disordered protein.[4] In 2010, scientists at the Brookhaven National Laboratory determined the X-ray crystal structure of the complex between Pup and its delivery enzyme Mpa 3M9D and found that Pup binding to Mpa induces the folding of a unique alpha-helix.[2]
In 2017, the presence of Pup homologs in bacterial species outside of the group of gram-positive bacteria was reported.[5] The Pup homologs were termed UBact (for Ubiquitin Bacterial), although the distinction has not been proven to be phylogenetically supported by a separate evolutionary origin and is without experimental evidence.[5] UBact is a homolog of Pup, and is found in several phyla of gram-negative bacteria (Pup is found predominantly in the gram-positive bacterial phylum Actinobacteria).
References
- Pearce, M. J.; Mintseris, J.; Ferreyra, J.; Gygi, S. P.; Darwin, K. H. (2008). "Ubiquitin-Like Protein Involved in the Proteasome Pathway of Mycobacterium tuberculosis". Science. 322 (5904): 1104–1107. doi:10.1126/science.1163885. PMC 2698935. PMID 18832610.
- Wang T, Darwin KH, Li H (November 2010). "Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation". Nature Structural & Molecular Biology. 17 (11): 1352–7. doi:10.1038/nsmb.1918. PMC 2988878. PMID 20953180.
- Universal protein resource accession number P9WHN4 for "Prokaryotic ubiquitin-like protein Pup" at UniProt.
- Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X (2009). "Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein". 422 (2). Biochemical Journal: 207–215. doi:10.1042/BJ20090738. PMID 19580545. Cite journal requires
|journal=(help) - Lehmann G, Udasin RG, Livneh I, Ciechanover A (February 2017). "Identification of UBact, a ubiquitin-like protein, along with other homologous components of a conjugation system and the proteasome in different gram-negative bacteria". Biochemical and Biophysical Research Communications. 483 (3): 946–950. doi:10.1016/j.bbrc.2017.01.037. PMID 28087277.
External links
- PupDB, a database of pupylated proteins and pupylation sites.