KCNJ12
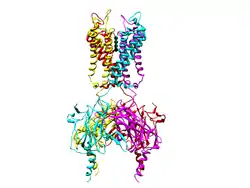
ATP-sensitive inward rectifier potassium channel 12 is a lipid-gated ion channel that in humans is encoded by the KCNJ12 gene.[5][6][7][8][9]
Function
This gene encodes an inwardly rectifying K+ channel that may be blocked by divalent cations. This protein is thought to be one of multiple inwardly rectifying channels that contribute to the cardiac inward rectifier current (IK1). The gene is located within the Smith–Magenis syndrome region on chromosome 17.[9]
Interactions
KCNJ12 has been shown to interact with:
See also
References
- GRCh38: Ensembl release 89: ENSG00000184185 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000042529 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Hansen, SB (May 2015). "Lipid agonism: The PIP2 paradigm of ligand-gated ion channels". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1851 (5): 620–8. doi:10.1016/j.bbalip.2015.01.011. PMC 4540326. PMID 25633344.
- Wible BA, De Biasi M, Majumder K, Taglialatela M, Brown AM (Mar 1995). "Cloning and functional expression of an inwardly rectifying K+ channel from human atrium". Circulation Research. 76 (3): 343–50. doi:10.1161/01.res.76.3.343. PMID 7859381.
- Kaibara M, Ishihara K, Doi Y, Hayashi H, Ehara T, Taniyama K (Nov 2002). "Identification of human Kir2.2 (KCNJ12) gene encoding functional inward rectifier potassium channel in both mammalian cells and Xenopus oocytes". FEBS Letters. 531 (2): 250–4. doi:10.1016/S0014-5793(02)03512-3. PMID 12417321. S2CID 46515689.
- Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, Lazdunski M, Nichols CG, Seino S, Vandenberg CA (Dec 2005). "International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels". Pharmacological Reviews. 57 (4): 509–26. doi:10.1124/pr.57.4.11. PMID 16382105. S2CID 11588492.
- "Entrez Gene: KCNJ12 potassium inwardly-rectifying channel, subfamily J, member 12".
- Leonoudakis D, Conti LR, Anderson S, Radeke CM, McGuire LM, Adams ME, Froehner SC, Yates JR, Vandenberg CA (May 2004). "Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins". The Journal of Biological Chemistry. 279 (21): 22331–46. doi:10.1074/jbc.M400285200. PMID 15024025.
- Leonoudakis D, Conti LR, Radeke CM, McGuire LM, Vandenberg CA (Apr 2004). "A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels". The Journal of Biological Chemistry. 279 (18): 19051–63. doi:10.1074/jbc.M400284200. PMID 14960569.
- Leonoudakis D, Mailliard W, Wingerd K, Clegg D, Vandenberg C (Mar 2001). "Inward rectifier potassium channel Kir2.2 is associated with synapse-associated protein SAP97". Journal of Cell Science. 114 (Pt 5): 987–98. PMID 11181181.
Further reading
- Namba N, Inagaki N, Gonoi T, Seino Y, Seino S (May 1996). "Kir2.2v: a possible negative regulator of the inwardly rectifying K+ channel Kir2.2". FEBS Letters. 386 (2–3): 211–4. doi:10.1016/0014-5793(96)00445-0. PMID 8647284. S2CID 44673403.
- Hugnot JP, Pedeutour F, Le Calvez C, Grosgeorge J, Passage E, Fontes M, Lazdunski M (Jan 1997). "The human inward rectifying K+ channel Kir 2.2 (KCNJ12) gene: gene structure, assignment to chromosome 17p11.1, and identification of a simple tandem repeat polymorphism". Genomics. 39 (1): 113–6. doi:10.1006/geno.1996.4450. PMID 9027495.
- Gallagher PG, Forget BG (Jan 1998). "An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene". The Journal of Biological Chemistry. 273 (3): 1339–48. doi:10.1074/jbc.273.3.1339. PMID 9430667.
- Namba N, Mori R, Tanaka H, Kondo I, Narahara K, Seino Y (1998). "The inwardly rectifying potassium channel subunit Kir2.2v (KCNJN1) maps to 17p11.2-->p11.1". Cytogenetics and Cell Genetics. 79 (1–2): 85–7. doi:10.1159/000134688. PMID 9533018.
- Leonoudakis D, Mailliard W, Wingerd K, Clegg D, Vandenberg C (Mar 2001). "Inward rectifier potassium channel Kir2.2 is associated with synapse-associated protein SAP97". Journal of Cell Science. 114 (Pt 5): 987–98. PMID 11181181.
- Preisig-Müller R, Schlichthörl G, Goerge T, Heinen S, Brüggemann A, Rajan S, Derst C, Veh RW, Daut J (May 2002). "Heteromerization of Kir2.x potassium channels contributes to the phenotype of Andersen's syndrome". Proceedings of the National Academy of Sciences of the United States of America. 99 (11): 7774–9. Bibcode:2002PNAS...99.7774P. doi:10.1073/pnas.102609499. PMC 124349. PMID 12032359.
- Chen L, Kawano T, Bajic S, Kaziro Y, Itoh H, Art JJ, Nakajima Y, Nakajima S (Jun 2002). "A glutamate residue at the C terminus regulates activity of inward rectifier K+ channels: implication for Andersen's syndrome". Proceedings of the National Academy of Sciences of the United States of America. 99 (12): 8430–5. Bibcode:2002PNAS...99.8430C. doi:10.1073/pnas.122682899. PMC 123084. PMID 12034888.
- Karkanis T, Li S, Pickering JG, Sims SM (Jun 2003). "Plasticity of KIR channels in human smooth muscle cells from internal thoracic artery". American Journal of Physiology. Heart and Circulatory Physiology. 284 (6): H2325–34. doi:10.1152/ajpheart.00559.2002. PMID 12598232.
- Stonehouse AH, Grubb BD, Pringle JH, Norman RI, Stanfield PR, Brammar WJ (Apr 2003). "Nuclear immunostaining in rat neuronal cells using two anti-Kir2.2 ion channel polyclonal antibodies". Journal of Molecular Neuroscience. 20 (2): 189–94. doi:10.1385/JMN:20:2:189. PMID 12794312. S2CID 34958415.
- Leonoudakis D, Conti LR, Radeke CM, McGuire LM, Vandenberg CA (Apr 2004). "A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels". The Journal of Biological Chemistry. 279 (18): 19051–63. doi:10.1074/jbc.M400284200. PMID 14960569.
- Leonoudakis D, Conti LR, Anderson S, Radeke CM, McGuire LM, Adams ME, Froehner SC, Yates JR, Vandenberg CA (May 2004). "Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins". The Journal of Biological Chemistry. 279 (21): 22331–46. doi:10.1074/jbc.M400285200. PMID 15024025.
- Fang Y, Schram G, Romanenko VG, Shi C, Conti L, Vandenberg CA, Davies PF, Nattel S, Levitan I (Nov 2005). "Functional expression of Kir2.x in human aortic endothelial cells: the dominant role of Kir2.2". American Journal of Physiology. Cell Physiology. 289 (5): C1134–44. doi:10.1152/ajpcell.00077.2005. PMID 15958527.
- Kiesecker C, Zitron E, Scherer D, Lueck S, Bloehs R, Scholz EP, Pirot M, Kathöfer S, Thomas D, Kreye VA, Kiehn J, Borst MM, Katus HA, Schoels W, Karle CA (Jan 2006). "Regulation of cardiac inwardly rectifying potassium current IK1 and Kir2.x channels by endothelin-1". Journal of Molecular Medicine. 84 (1): 46–56. doi:10.1007/s00109-005-0707-8. PMID 16258766. S2CID 2081384.
External links
- Kir2.2+channel at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.